Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod99
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod99 |
| Module size |
39 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 8086
|
AAAS
|
aladin WD repeat nucleoporin
|
| 11183
|
GCKR
|
mitogen-activated protein kinase kinase kinase kinase 5
|
| 8487
|
GEMIN2
|
gem nuclear organelle associated protein 2
|
| 50628
|
GEMIN4
|
gem nuclear organelle associated protein 4
|
| 25929
|
GEMIN5
|
gem nuclear organelle associated protein 5
|
| 79833
|
GEMIN6
|
gem nuclear organelle associated protein 6
|
| 79760
|
GEMIN7
|
gem nuclear organelle associated protein 7
|
| 54960
|
GEMIN8
|
gem nuclear organelle associated protein 8
|
| 500697
|
GLE1
|
similar to GLE1-like, RNA export mediator isoform 1
|
| 57727
|
NCOA5
|
nuclear receptor coactivator 5
|
| 91754
|
NEK9
|
NIMA related kinase 9
|
| 9972
|
NUP153
|
nucleoporin 153
|
| 9631
|
NUP155
|
nucleoporin 155
|
| 23511
|
NUP188
|
nucleoporin 188
|
| 23165
|
NUP205
|
nucleoporin 205
|
| 23225
|
NUP210
|
nucleoporin 210
|
| 8021
|
NUP214
|
nucleoporin 214
|
| 129401
|
NUP35
|
nucleoporin 35
|
| 25497
|
NUP50
|
nucleoporin 50
|
| 53371
|
NUP54
|
nucleoporin 54
|
| 23636
|
NUP62
|
nucleoporin 62
|
| 4927
|
NUP88
|
nucleoporin 88
|
| 9688
|
NUP93
|
nucleoporin 93
|
| 4928
|
NUP98
|
nucleoporin 98
|
| 9818
|
NUPL1
|
nucleoporin 58
|
| 11097
|
NUPL2
|
nucleoporin like 2
|
| 10482
|
NXF1
|
nuclear RNA export factor 1
|
| 56001
|
NXF2
|
nuclear RNA export factor 2
|
| 728343
|
NXF2B
|
nuclear RNA export factor 2B
|
| 56000
|
NXF3
|
nuclear RNA export factor 3
|
| 55998
|
NXF5
|
nuclear RNA export factor 5
|
| 29107
|
NXT1
|
nuclear transport factor 2 like export factor 1
|
| 55916
|
NXT2
|
nuclear transport factor 2 like export factor 2
|
| 51808
|
PHAX
|
phosphorylated adaptor for RNA export
|
| 8480
|
RAE1
|
ribonucleic acid export 1
|
| 126638
|
RPTN
|
repetin
|
| 7884
|
SLBP
|
stem-loop binding protein
|
| 10073
|
SNUPN
|
snurportin 1
|
| 7175
|
TPR
|
translocated promoter region, nuclear basket protein
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
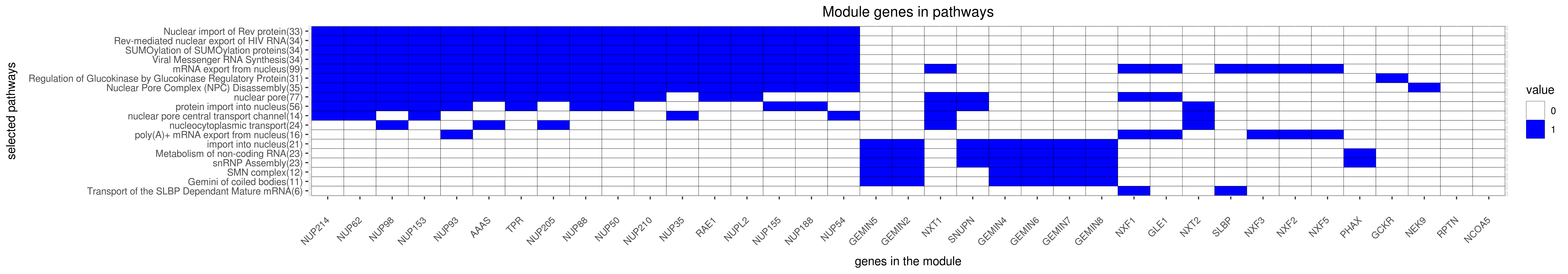
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0e+00
|
0e+00
|
PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA
|
| 0e+00
|
0e+00
|
MRNA PROCESSING
|
| 0e+00
|
0e+00
|
TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM
|
| 0e+00
|
0e+00
|
METABOLISM OF NON CODING RNA
|
| 0e+00
|
0e+00
|
SLC MEDIATED TRANSMEMBRANE TRANSPORT
|
| 0e+00
|
0e+00
|
REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN
|
| 0e+00
|
0e+00
|
INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS
|
| 0e+00
|
0e+00
|
METABOLISM OF CARBOHYDRATES
|
| 0e+00
|
0e+00
|
NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY
|
| 0e+00
|
0e+00
|
GLUCOSE TRANSPORT
|
| 0e+00
|
0e+00
|
ANTIVIRAL MECHANISM BY IFN STIMULATED GENES
|
| 0e+00
|
0e+00
|
TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES
|
| 0e+00
|
0e+00
|
INFLUENZA LIFE CYCLE
|
| 0e+00
|
0e+00
|
TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT
|
| 0e+00
|
0e+00
|
HIV LIFE CYCLE
|
| 0e+00
|
0e+00
|
LATE PHASE OF HIV LIFE CYCLE
|
| 0e+00
|
0e+00
|
METABOLISM OF RNA
|
| 0e+00
|
0e+00
|
INTERFERON SIGNALING
|
| 0e+00
|
0e+00
|
HOST INTERACTIONS OF HIV FACTORS
|
| 0e+00
|
0e+00
|
TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:24:36 2018 - R2HTML