Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod85
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod85 |
| Module size |
57 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 3983
|
ABLIM1
|
actin binding LIM protein 1
|
| 84448
|
ABLIM2
|
actin binding LIM protein family member 2
|
| 22885
|
ABLIM3
|
actin binding LIM protein family member 3
|
| 8851
|
CDK5R1
|
cyclin dependent kinase 5 regulatory subunit 1
|
| 1630
|
DCC
|
DCC netrin 1 receptor
|
| 1826
|
DSCAM
|
DS cell adhesion molecule
|
| 57453
|
DSCAML1
|
DS cell adhesion molecule like 1
|
| 9855
|
FARP2
|
FERM, ARH/RhoGEF and pleckstrin domain protein 2
|
| 2242
|
FES
|
FES proto-oncogene, tyrosine kinase
|
| 84624
|
FNDC1
|
fibronectin type III domain containing 1
|
| 26762
|
HAVCR1
|
hepatitis A virus cellular receptor 1
|
| 9543
|
IGDCC3
|
immunoglobulin superfamily DCC subclass member 3
|
| 57722
|
IGDCC4
|
immunoglobulin superfamily DCC subclass member 4
|
| 57214
|
KIAA1199
|
cell migration inducing hyaluronidase 1
|
| 148930
|
KNCN
|
kinocilin
|
| 4756
|
NEO1
|
neogenin 1
|
| 4745
|
NRP1
|
neural EGFL like 1
|
| 4753
|
NRP2
|
neural EGFL like 2
|
| 9423
|
NTN1
|
netrin 1
|
| 4917
|
NTN3
|
netrin 3
|
| 126147
|
NTN5
|
netrin 5
|
| 84628
|
NTNG1
|
netrin G2
|
| 84628
|
NTNG2
|
netrin G2
|
| 5361
|
PLXNA1
|
plexin A1
|
| 5362
|
PLXNA2
|
plexin A2
|
| 55558
|
PLXNA3
|
plexin A3
|
| 91584
|
PLXNA4
|
plexin A4
|
| 5364
|
PLXNB1
|
plexin B1
|
| 23654
|
PLXNB2
|
plexin B2
|
| 10154
|
PLXNC1
|
plexin C1
|
| 23129
|
PLXND1
|
plexin D1
|
| 283659
|
PRTG
|
protogenin
|
| 10371
|
SEMA3A
|
semaphorin 3A
|
| 10512
|
SEMA3C
|
semaphorin 3C
|
| 223117
|
SEMA3D
|
semaphorin 3D
|
| 9723
|
SEMA3E
|
semaphorin 3E
|
| 6405
|
SEMA3F
|
semaphorin 3F
|
| 56920
|
SEMA3G
|
semaphorin 3G
|
| 64218
|
SEMA4A
|
semaphorin 4A
|
| 10509
|
SEMA4B
|
semaphorin 4B
|
| 54910
|
SEMA4C
|
semaphorin 4C
|
| 10505
|
SEMA4F
|
ssemaphorin 4F
|
| 57715
|
SEMA4G
|
semaphorin 4G
|
| 9037
|
SEMA5A
|
semaphorin 5A
|
| 54437
|
SEMA5B
|
semaphorin 5B
|
| 57556
|
SEMA6A
|
semaphorin 6A
|
| 10501
|
SEMA6B
|
semaphorin 6B
|
| 10500
|
SEMA6C
|
semaphorin 6C
|
| 80031
|
SEMA6D
|
semaphorin 6D
|
| 8482
|
SEMA7A
|
semaphorin 7A (John Milton Hagen blood group)
|
| 80723
|
SLC35G2
|
solute carrier family 35 member G2
|
| 440730
|
TRIM67
|
tripartite motif containing 67
|
| 90249
|
UNC5A
|
unc-5 netrin receptor A
|
| 219699
|
UNC5B
|
unc-5 netrin receptor B
|
| 8633
|
UNC5C
|
unc-5 netrin receptor C
|
| 222643
|
UNC5CL
|
unc-5 family C-terminal like
|
| 137970
|
UNC5D
|
unc-5 netrin receptor D
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
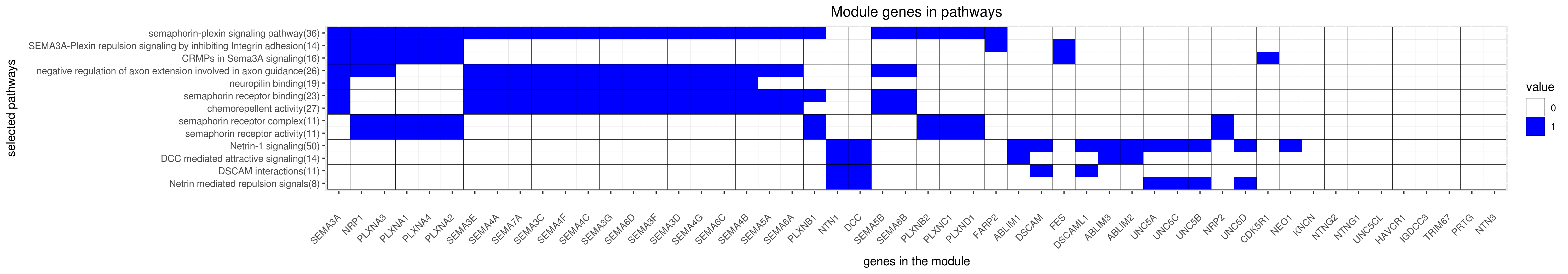
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0.00e+00
|
0.00e+00
|
AXON GUIDANCE
|
| 0.00e+00
|
0.00e+00
|
SEMAPHORIN INTERACTIONS
|
| 0.00e+00
|
0.00e+00
|
OTHER SEMAPHORIN INTERACTIONS
|
| 0.00e+00
|
0.00e+00
|
DEVELOPMENTAL BIOLOGY
|
| 0.00e+00
|
0.00e+00
|
NETRIN1 SIGNALING
|
| 0.00e+00
|
0.00e+00
|
CRMPS IN SEMA3A SIGNALING
|
| 0.00e+00
|
0.00e+00
|
SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION
|
| 0.00e+00
|
0.00e+00
|
AXON GUIDANCE
|
| 0.00e+00
|
0.00e+00
|
SEMAPHORIN INTERACTIONS
|
| 0.00e+00
|
0.00e+00
|
OTHER SEMAPHORIN INTERACTIONS
|
| 0.00e+00
|
0.00e+00
|
NETRIN1 SIGNALING
|
| 5.37e-12
|
2.08e-09
|
SEMA3A PAK DEPENDENT AXON REPULSION
|
| 1.20e-11
|
3.50e-09
|
SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION
|
| 9.61e-11
|
2.63e-08
|
CRMPS IN SEMA3A SIGNALING
|
| 1.25e-09
|
3.01e-07
|
SEMA3A PAK DEPENDENT AXON REPULSION
|
| 1.32e-08
|
3.81e-06
|
DSCAM INTERACTIONS
|
| 2.09e-08
|
4.42e-06
|
DSCAM INTERACTIONS
|
| 8.11e-08
|
2.11e-05
|
DCC MEDIATED ATTRACTIVE SIGNALING
|
| 2.84e-07
|
5.30e-05
|
DCC MEDIATED ATTRACTIVE SIGNALING
|
| 4.55e-05
|
7.68e-03
|
ROLE OF DCC IN REGULATING APOPTOSIS
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:23:54 2018 - R2HTML