Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod76
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod76 |
| Module size |
22 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 8708
|
B3GALT1
|
beta-1,3-galactosyltransferase 1
|
| 8707
|
B3GALT2
|
beta-1,3-galactosyltransferase 2
|
| 10317
|
B3GALT5
|
beta-1,3-galactosyltransferase 5
|
| 10678
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2
|
| 2683
|
B4GALT1
|
beta-1,4-galactosyltransferase 1
|
| 8704
|
B4GALT2
|
beta-1,4-galactosyltransferase 2
|
| 8703
|
B4GALT3
|
beta-1,4-galactosyltransferase 3
|
| 8702
|
B4GALT4
|
beta-1,4-galactosyltransferase 4
|
| 2523
|
FUT1
|
fucosyltransferase 1 (H blood group)
|
| 2524
|
FUT2
|
fucosyltransferase 2
|
| 2525
|
FUT3
|
fucosyltransferase 3 (Lewis blood group)
|
| 2526
|
FUT4
|
fucosyltransferase 4
|
| 2527
|
FUT5
|
fucosyltransferase 5
|
| 2528
|
FUT6
|
fucosyltransferase 6
|
| 2529
|
FUT7
|
fucosyltransferase 7
|
| 10690
|
FUT9
|
fucosyltransferase 9
|
| 2651
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group)
|
| 120071
|
GYLTL1B
|
LARGE xylosyl- and glucuronyltransferase 2
|
| 9215
|
LARGE
|
LARGE xylosyl- and glucuronyltransferase 1
|
| 10402
|
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6
|
| 84620
|
ST6GAL2
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 2
|
| 6489
|
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
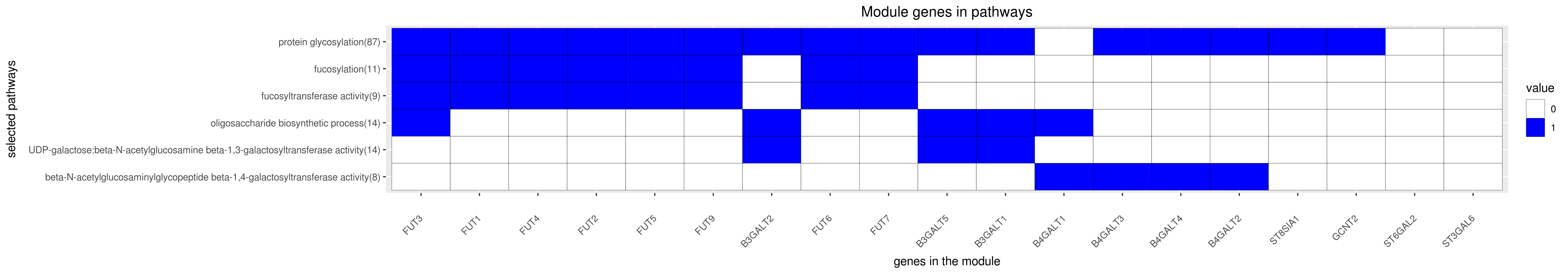
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0.00e+00
|
0.00e+00
|
KERATAN SULFATE BIOSYNTHESIS
|
| 3.08e-12
|
1.20e-09
|
KERATAN SULFATE BIOSYNTHESIS
|
| 7.76e-12
|
2.27e-09
|
KERATAN SULFATE KERATIN METABOLISM
|
| 3.22e-11
|
1.21e-08
|
KERATAN SULFATE KERATIN METABOLISM
|
| 9.69e-09
|
2.14e-06
|
GLYCOSAMINOGLYCAN METABOLISM
|
| 1.34e-08
|
3.86e-06
|
N GLYCAN ANTENNAE ELONGATION
|
| 1.49e-08
|
4.27e-06
|
GLYCOSAMINOGLYCAN METABOLISM
|
| 2.33e-08
|
6.55e-06
|
N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI
|
| 5.30e-08
|
1.07e-05
|
N GLYCAN ANTENNAE ELONGATION
|
| 8.47e-08
|
1.67e-05
|
METABOLISM OF CARBOHYDRATES
|
| 9.40e-08
|
1.84e-05
|
N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI
|
| 1.76e-07
|
4.43e-05
|
TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION
|
| 4.43e-07
|
1.05e-04
|
METABOLISM OF CARBOHYDRATES
|
| 1.65e-06
|
2.77e-04
|
TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION
|
| 1.67e-06
|
3.61e-04
|
ASPARAGINE N LINKED GLYCOSYLATION
|
| 8.34e-06
|
1.26e-03
|
ASPARAGINE N LINKED GLYCOSYLATION
|
| 1.89e-05
|
2.71e-03
|
POST TRANSLATIONAL PROTEIN MODIFICATION
|
| 5.26e-05
|
8.74e-03
|
POST TRANSLATIONAL PROTEIN MODIFICATION
|
| 4.78e-04
|
6.18e-02
|
PRE NOTCH PROCESSING IN GOLGI
|
| 1.42e-03
|
1.37e-01
|
PRE NOTCH PROCESSING IN GOLGI
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:23:27 2018 - R2HTML