Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod7
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod7 |
| Module size |
89 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 140690
|
CTCFL
|
CCCTC-binding factor like
|
| 6018
|
RLF
|
rearranged L-myc fusion
|
| 65986
|
ZBTB10
|
zinc finger and BTB domain containing 10
|
| 57621
|
ZBTB2
|
zinc finger and BTB domain containing 2
|
| 10009
|
ZBTB33
|
zinc finger and BTB domain containing 33
|
| 403341
|
ZBTB34
|
zinc finger and BTB domain containing 34
|
| 80108
|
ZFP2
|
ZFP2 zinc finger protein
|
| 139735
|
ZFP92
|
ZFP92 zinc finger protein
|
| 284307
|
ZIK1
|
zinc finger protein interacting with K protein 1
|
| 163227
|
ZNF100
|
zinc finger protein 100
|
| 51427
|
ZNF107
|
zinc finger protein 107
|
| 7692
|
ZNF133
|
zinc finger protein 133
|
| 7699
|
ZNF140
|
zinc finger protein 140
|
| 7700
|
ZNF141
|
zinc finger protein 141
|
| 7705
|
ZNF146
|
zinc finger protein 146
|
| 7711
|
ZNF155
|
zinc finger protein 155
|
| 7712
|
ZNF157
|
zinc finger protein 157
|
| 90338
|
ZNF160
|
zinc finger protein 160
|
| 7743
|
ZNF189
|
zinc finger protein 189
|
| 7752
|
ZNF200
|
zinc finger protein 200
|
| 7988
|
ZNF212
|
zinc finger protein 212
|
| 7761
|
ZNF214
|
zinc finger protein 214
|
| 7770
|
ZNF227
|
zinc finger protein 227
|
| 7551
|
ZNF3
|
zinc finger protein 3
|
| 57343
|
ZNF304
|
zinc finger protein 304
|
| 150
|
ZNF32
|
adrenoceptor alpha 2A
|
| 79175
|
ZNF343
|
zinc finger protein 343
|
| 7584
|
ZNF35
|
zinc finger protein 35
|
| 57541
|
ZNF398
|
zinc finger protein 398
|
| 84330
|
ZNF414
|
zinc finger protein 414
|
| 55786
|
ZNF415
|
zinc finger protein 415
|
| 79088
|
ZNF426
|
zinc finger protein 426
|
| 353088
|
ZNF429
|
zinc finger protein 429
|
| 220929
|
ZNF438
|
zinc finger protein 438
|
| 126070
|
ZNF440
|
zinc finger protein 440
|
| 388566
|
ZNF470
|
zinc finger protein 470
|
| 220992
|
ZNF485
|
zinc finger protein 485
|
| 26048
|
ZNF500
|
zinc finger protein 500
|
| 440515
|
ZNF506
|
zinc finger protein 506
|
| 84450
|
ZNF512
|
zinc finger protein 512
|
| 9658
|
ZNF516
|
zinc finger protein 516
|
| 340385
|
ZNF517
|
zinc finger protein 517
|
| 348327
|
ZNF530
|
zinc finger protein 530
|
| 284306
|
ZNF547
|
zinc finger protein 547
|
| 79818
|
ZNF552
|
zinc finger protein 552
|
| 84527
|
ZNF559
|
zinc finger protein 559
|
| 100529215
|
ZNF559-ZNF177
|
ZNF559-ZNF177 readthrough
|
| 126295
|
ZNF57
|
zinc finger protein 57
|
| 163033
|
ZNF579
|
zinc finger protein 579
|
| 84914
|
ZNF587
|
zinc finger protein 587
|
| 152687
|
ZNF595
|
zinc finger protein 595
|
| 169270
|
ZNF596
|
zinc finger protein 596
|
| 148103
|
ZNF599
|
zinc finger protein 599
|
| 84775
|
ZNF607
|
zinc finger protein 607
|
| 90317
|
ZNF616
|
zinc finger protein 616
|
| 57547
|
ZNF624
|
zinc finger protein 624
|
| 90589
|
ZNF625
|
zinc finger protein 625
|
| 51193
|
ZNF639
|
zinc finger protein 639
|
| 9726
|
ZNF646
|
zinc finger protein 646
|
| 55279
|
ZNF654
|
zinc finger protein 654
|
| 79027
|
ZNF655
|
zinc finger protein 655
|
| 285349
|
ZNF660
|
zinc finger protein 660
|
| 79894
|
ZNF672
|
zinc finger protein 672
|
| 641339
|
ZNF674
|
zinc finger protein 674
|
| 168417
|
ZNF679
|
zinc finger protein 679
|
| 340252
|
ZNF680
|
zinc finger protein 680
|
| 127396
|
ZNF684
|
zinc finger protein 684
|
| 146542
|
ZNF688
|
zinc finger protein 688
|
| 55657
|
ZNF692
|
zinc finger protein 692
|
| 124411
|
ZNF720
|
zinc finger protein 720
|
| 170960
|
ZNF721
|
zinc finger protein 721
|
| 388523
|
ZNF728
|
zinc finger protein 728
|
| 148203
|
ZNF738
|
zinc finger protein 738
|
| 7625
|
ZNF74
|
zinc finger protein 74
|
| 7627
|
ZNF75A
|
zinc finger protein 75a
|
| 91661
|
ZNF765
|
zinc finger protein 765
|
| 79724
|
ZNF768
|
zinc finger protein 768
|
| 58492
|
ZNF77
|
zinc finger protein 77
|
| 284323
|
ZNF780A
|
zinc finger protein 780A
|
| 136051
|
ZNF786
|
zinc finger protein 786
|
| 388536
|
ZNF790
|
zinc finger protein 790
|
| 126375
|
ZNF792
|
zinc finger protein 792
|
| 390980
|
ZNF805
|
zinc finger protein 805
|
| 388558
|
ZNF808
|
zinc finger protein 808
|
| 55769
|
ZNF83
|
zinc finger protein 83
|
| 7637
|
ZNF84
|
zinc finger protein 84
|
| 7639
|
ZNF85
|
zinc finger protein 85
|
| 168374
|
ZNF92
|
zinc finger protein 92
|
| 222696
|
ZSCAN23
|
zinc finger and SCAN domain containing 23
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
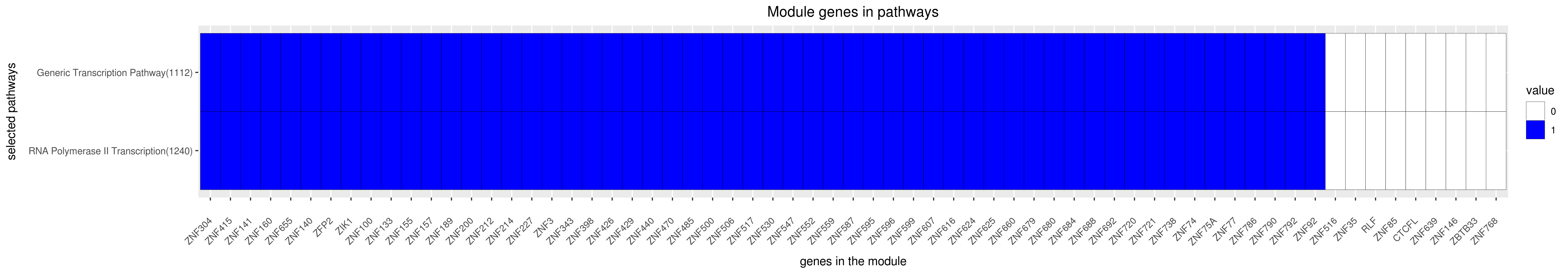
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0e+00
|
0e+00
|
GENERIC TRANSCRIPTION PATHWAY
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:19:58 2018 - R2HTML