Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod62
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod62 |
| Module size |
18 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 6529
|
SLC6A1
|
solute carrier family 6 member 1
|
| 6538
|
SLC6A11
|
solute carrier family 6 member 11
|
| 6539
|
SLC6A12
|
solute carrier family 6 member 12
|
| 6540
|
SLC6A13
|
solute carrier family 6 member 13
|
| 11254
|
SLC6A14
|
solute carrier family 6 member 14
|
| 55117
|
SLC6A15
|
solute carrier family 6 member 15
|
| 388662
|
SLC6A17
|
solute carrier family 6 member 17
|
| 348932
|
SLC6A18
|
solute carrier family 6 member 18
|
| 340024
|
SLC6A19
|
solute carrier family 6 member 19
|
| 6530
|
SLC6A2
|
solute carrier family 6 member 2
|
| 54716
|
SLC6A20
|
solute carrier family 6 member 20
|
| 6532
|
SLC6A4
|
solute carrier family 6 member 4
|
| 9152
|
SLC6A5
|
solute carrier family 6 member 5
|
| 6533
|
SLC6A6
|
solute carrier family 6 member 6
|
| 6534
|
SLC6A7
|
solute carrier family 6 member 7
|
| 6535
|
SLC6A8
|
solute carrier family 6 member 8
|
| 6536
|
SLC6A9
|
solute carrier family 6 member 9
|
| 57393
|
TMEM27
|
collectrin, amino acid transport regulator
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
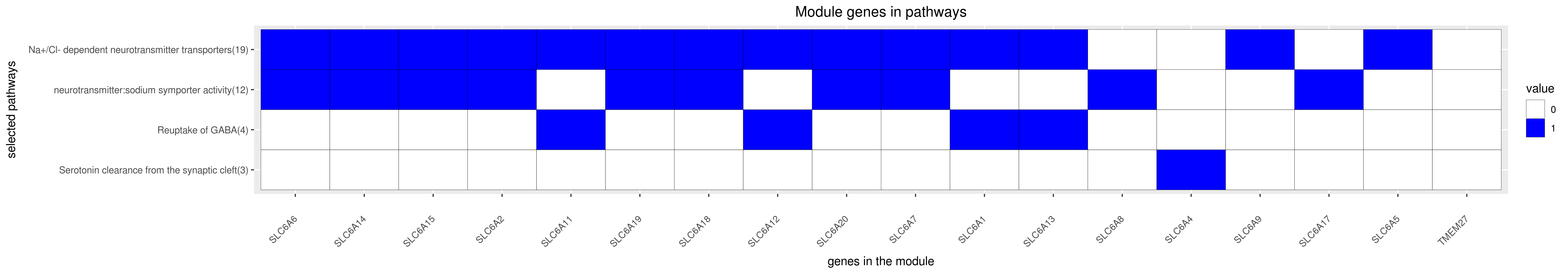
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0.00e+00
|
0.00e+00
|
TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES
|
| 0.00e+00
|
0.00e+00
|
NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS
|
| 0.00e+00
|
0.00e+00
|
SLC MEDIATED TRANSMEMBRANE TRANSPORT
|
| 0.00e+00
|
0.00e+00
|
AMINE COMPOUND SLC TRANSPORTERS
|
| 0.00e+00
|
0.00e+00
|
TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS
|
| 0.00e+00
|
0.00e+00
|
NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS
|
| 0.00e+00
|
0.00e+00
|
SLC MEDIATED TRANSMEMBRANE TRANSPORT
|
| 0.00e+00
|
0.00e+00
|
AMINE COMPOUND SLC TRANSPORTERS
|
| 0.00e+00
|
0.00e+00
|
TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS
|
| 1.04e-10
|
2.84e-08
|
AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE
|
| 2.51e-10
|
8.78e-08
|
AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE
|
| 1.99e-09
|
4.67e-07
|
AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS
|
| 2.61e-09
|
8.11e-07
|
AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS
|
| 3.75e-09
|
1.14e-06
|
TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES
|
| 4.10e-09
|
9.35e-07
|
TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES
|
| 1.80e-05
|
3.28e-03
|
GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION
|
| 2.80e-04
|
3.86e-02
|
NEUROTRANSMITTER RELEASE CYCLE
|
| 2.90e-04
|
3.33e-02
|
GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION
|
| 2.61e-03
|
2.35e-01
|
NEUROTRANSMITTER RELEASE CYCLE
|
| 1.23e-02
|
9.39e-01
|
TRANSMISSION ACROSS CHEMICAL SYNAPSES
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:22:44 2018 - R2HTML