Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod54
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod54 |
| Module size |
52 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 22794
|
CASC3
|
CASC3, exon junction complex subunit
|
| 4736
|
RPL10A
|
ribosomal protein L10a
|
| 6136
|
RPL12
|
ribosomal protein L12
|
| 28998
|
RPL13
|
mitochondrial ribosomal protein L13
|
| 9045
|
RPL14
|
ribosomal protein L14
|
| 6141
|
RPL18
|
ribosomal protein L18
|
| 6142
|
RPL18A
|
ribosomal protein L18a
|
| 6143
|
RPL19
|
ribosomal protein L19
|
| 6144
|
RPL21
|
ribosomal protein L21
|
| 6146
|
RPL22
|
ribosomal protein L22
|
| 6147
|
RPL23A
|
ribosomal protein L23a
|
| 51187
|
RPL24
|
ribosomal L24 domain containing 1
|
| 6154
|
RPL26
|
ribosomal protein L26
|
| 51121
|
RPL26L1
|
ribosomal protein L26 like 1
|
| 6155
|
RPL27
|
ribosomal protein L27
|
| 6158
|
RPL28
|
ribosomal protein L28
|
| 6159
|
RPL29
|
ribosomal protein L29
|
| 6156
|
RPL30
|
ribosomal protein L30
|
| 6160
|
RPL31
|
ribosomal protein L31
|
| 6161
|
RPL32
|
ribosomal protein L32
|
| 6164
|
RPL34
|
ribosomal protein L34
|
| 6165
|
RPL35A
|
ribosomal protein L35a
|
| 25873
|
RPL36
|
ribosomal protein L36
|
| 6167
|
RPL37
|
ribosomal protein L37
|
| 6168
|
RPL37A
|
ribosomal protein L37a
|
| 6169
|
RPL38
|
ribosomal protein L38
|
| 6170
|
RPL39
|
ribosomal protein L39
|
| 6171
|
RPL41
|
ribosomal protein L41
|
| 6128
|
RPL6
|
ribosomal protein L6
|
| 6175
|
RPLP0
|
ribosomal protein lateral stalk subunit P0
|
| 6176
|
RPLP1
|
ribosomal protein lateral stalk subunit P1
|
| 6181
|
RPLP2
|
ribosomal protein lateral stalk subunit P2
|
| 6204
|
RPS10
|
ribosomal protein S10
|
| 6183
|
RPS12
|
mitochondrial ribosomal protein S12
|
| 6208
|
RPS14
|
ribosomal protein S14
|
| 6210
|
RPS15A
|
ribosomal protein S15a
|
| 6223
|
RPS19
|
ribosomal protein S19
|
| 6227
|
RPS21
|
ribosomal protein S21
|
| 6229
|
RPS24
|
ribosomal protein S24
|
| 6231
|
RPS26
|
ribosomal protein S26
|
| 6232
|
RPS27
|
ribosomal protein S27
|
| 6234
|
RPS28
|
ribosomal protein S28
|
| 6235
|
RPS29
|
ribosomal protein S29
|
| 6189
|
RPS3A
|
ribosomal protein S3A
|
| 6191
|
RPS4X
|
ribosomal protein S4 X-linked
|
| 6192
|
RPS4Y1
|
ribosomal protein S4 Y-linked 1
|
| 6201
|
RPS7
|
ribosomal protein S7
|
| 23381
|
SMG5
|
SMG5, nonsense mediated mRNA decay factor
|
| 23293
|
SMG6
|
SMG6, nonsense mediated mRNA decay factor
|
| 9887
|
SMG7
|
SMG7, nonsense mediated mRNA decay factor
|
| 55181
|
SMG8
|
SMG8, nonsense mediated mRNA decay factor
|
| 56006
|
SMG9
|
SMG9, nonsense mediated mRNA decay factor
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
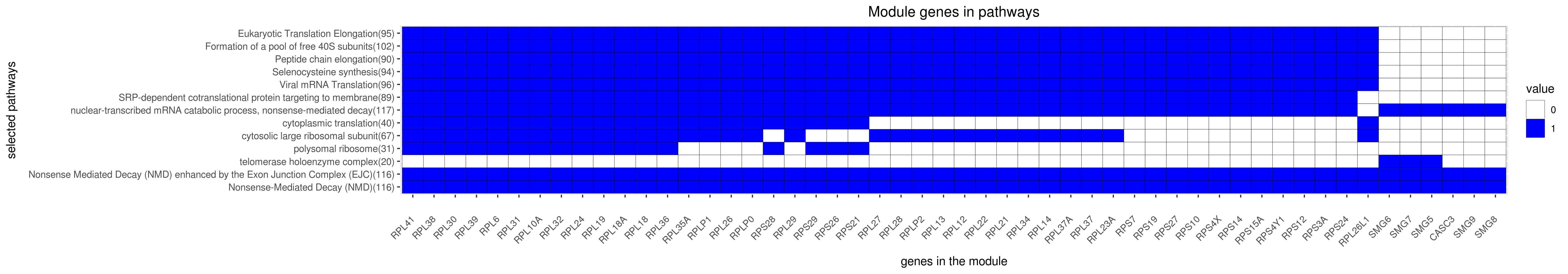
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0e+00
|
0e+00
|
INFLUENZA LIFE CYCLE
|
| 0e+00
|
0e+00
|
INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION
|
| 0e+00
|
0e+00
|
METABOLISM OF RNA
|
| 0e+00
|
0e+00
|
SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE
|
| 0e+00
|
0e+00
|
PEPTIDE CHAIN ELONGATION
|
| 0e+00
|
0e+00
|
NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX
|
| 0e+00
|
0e+00
|
METABOLISM OF MRNA
|
| 0e+00
|
0e+00
|
TRANSLATION
|
| 0e+00
|
0e+00
|
3 UTR MEDIATED TRANSLATIONAL REGULATION
|
| 0e+00
|
0e+00
|
ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S
|
| 0e+00
|
0e+00
|
FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX
|
| 0e+00
|
0e+00
|
INFLUENZA LIFE CYCLE
|
| 0e+00
|
0e+00
|
INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION
|
| 0e+00
|
0e+00
|
METABOLISM OF RNA
|
| 0e+00
|
0e+00
|
SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE
|
| 0e+00
|
0e+00
|
PEPTIDE CHAIN ELONGATION
|
| 0e+00
|
0e+00
|
NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX
|
| 0e+00
|
0e+00
|
METABOLISM OF MRNA
|
| 0e+00
|
0e+00
|
3 UTR MEDIATED TRANSLATIONAL REGULATION
|
| 0e+00
|
0e+00
|
TRANSLATION
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:22:21 2018 - R2HTML