Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod38
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod38 |
| Module size |
61 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 1747
|
DLX3
|
distal-less homeobox 3
|
| 2128
|
EVX1
|
even-skipped homeobox 1
|
| 344191
|
EVX2
|
even-skipped homeobox 2
|
| 2636
|
GBX1
|
gastrulation brain homeobox 1
|
| 2637
|
GBX2
|
gastrulation brain homeobox 2
|
| 219409
|
GSX1
|
GS homeobox 1
|
| 170825
|
GSX2
|
GS homeobox 2
|
| 3206
|
HOXA10
|
homeobox A10
|
| 3207
|
HOXA11
|
homeobox A11
|
| 3209
|
HOXA13
|
homeobox A13
|
| 3199
|
HOXA2
|
homeobox A2
|
| 3200
|
HOXA3
|
homeobox A3
|
| 3201
|
HOXA4
|
homeobox A4
|
| 3202
|
HOXA5
|
homeobox A5
|
| 3211
|
HOXB1
|
homeobox B1
|
| 10481
|
HOXB13
|
homeobox B13
|
| 3212
|
HOXB2
|
homeobox B2
|
| 3213
|
HOXB3
|
homeobox B3
|
| 3214
|
HOXB4
|
homeobox B4
|
| 3215
|
HOXB5
|
homeobox B5
|
| 3216
|
HOXB6
|
homeobox B6
|
| 3217
|
HOXB7
|
homeobox B7
|
| 3218
|
HOXB8
|
homeobox B8
|
| 3219
|
HOXB9
|
homeobox B9
|
| 3226
|
HOXC10
|
homeobox C10
|
| 3227
|
HOXC11
|
homeobox C11
|
| 3228
|
HOXC12
|
homeobox C12
|
| 3229
|
HOXC13
|
homeobox C13
|
| 3221
|
HOXC4
|
homeobox C4
|
| 3222
|
HOXC5
|
homeobox C5
|
| 3223
|
HOXC6
|
homeobox C6
|
| 3224
|
HOXC8
|
homeobox C8
|
| 3225
|
HOXC9
|
homeobox C9
|
| 3231
|
HOXD1
|
homeobox D1
|
| 3236
|
HOXD10
|
homeobox D10
|
| 3237
|
HOXD11
|
homeobox D11
|
| 3238
|
HOXD12
|
homeobox D12
|
| 3239
|
HOXD13
|
homeobox D13
|
| 3232
|
HOXD3
|
homeobox D3
|
| 3233
|
HOXD4
|
homeobox D4
|
| 3234
|
HOXD8
|
homeobox D8
|
| 3235
|
HOXD9
|
homeobox D9
|
| 79192
|
IRX1
|
iroquois homeobox 1
|
| 153572
|
IRX2
|
iroquois homeobox 2
|
| 10265
|
IRX5
|
iroquois homeobox 5
|
| 55180
|
LINS
|
lines homolog 1
|
| 4211
|
MEIS1
|
Meis homeobox 1
|
| 4212
|
MEIS2
|
Meis homeobox 2
|
| 56917
|
MEIS3
|
Meis homeobox 3
|
| 4222
|
MEOX1
|
mesenchyme homeobox 1
|
| 3110
|
MNX1
|
motor neuron and pancreas homeobox 1
|
| 5087
|
PBX1
|
PBX homeobox 1
|
| 5089
|
PBX2
|
PBX homeobox 2
|
| 5090
|
PBX3
|
PBX homeobox 3
|
| 80714
|
PBX4
|
PBX homeobox 4
|
| 267004
|
PGBD3
|
piggyBac transposable element derived 3
|
| 10745
|
PHTF1
|
putative homeodomain transcription factor 1
|
| 5316
|
PKNOX1
|
PBX/knotted 1 homeobox 1
|
| 90316
|
TGIF2LX
|
TGFB induced factor homeobox 2 like X-linked
|
| 90655
|
TGIF2LY
|
TGFB induced factor homeobox 2 like Y-linked
|
| 3195
|
TLX1
|
T cell leukemia homeobox 1
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
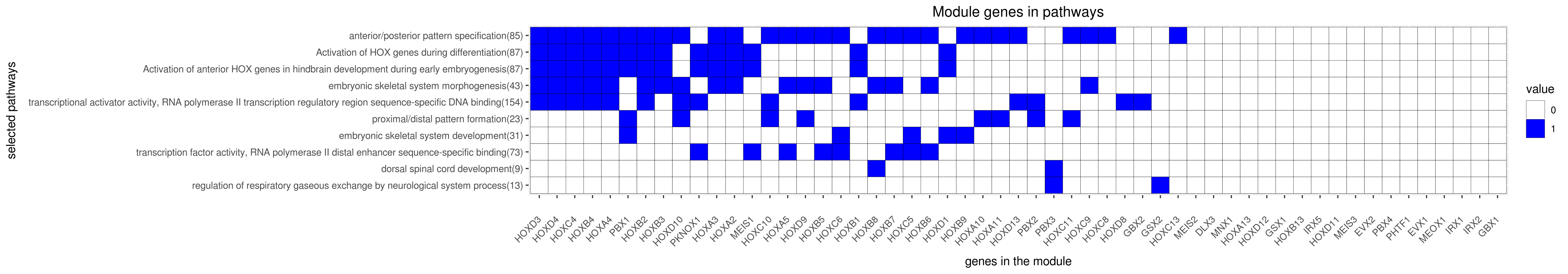
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:21:32 2018 - R2HTML