Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod376
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod376 |
| Module size |
32 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 27236
|
ARFIP1
|
ADP ribosylation factor interacting protein 1
|
| 23621
|
BACE1
|
beta-secretase 1
|
| 8678
|
BECN1
|
beclin 1
|
| 79839
|
CCDC102B
|
coiled-coil domain containing 102B
|
| 80157
|
CWH43
|
cell wall biogenesis 43 C-terminal homolog
|
| 79901
|
CYBRD1
|
cytochrome b reductase 1
|
| 23259
|
DDHD2
|
DDHD domain containing 2
|
| 10059
|
DNM1L
|
dynamin 1 like
|
| 79180
|
EFHD2
|
EF-hand domain family member D2
|
| 57795
|
FAM5B
|
BMP/retinoic acid inducible neural specific 2
|
| 54874
|
FNBP1L
|
formin binding protein 1 like
|
| 9648
|
GCC2
|
GRIP and coiled-coil domain containing 2
|
| 84514
|
GHDC
|
GH3 domain containing
|
| 2764
|
GMFB
|
glia maturation factor beta
|
| 64083
|
GOLPH3
|
golgi phosphoprotein 3
|
| 2822
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1
|
| 55196
|
KIAA1551
|
KIAA1551
|
| 3881
|
KRT31
|
keratin 31
|
| 3885
|
KRT34
|
keratin 34
|
| 3916
|
LAMP1
|
lysosomal associated membrane protein 1
|
| 83876
|
MRO
|
maestro
|
| 4599
|
MX1
|
MX dynamin like GTPase 1
|
| 27315
|
PGAP2
|
post-GPI attachment to proteins 2
|
| 6271
|
S100A1
|
S100 calcium binding protein A1
|
| 80851
|
SH3BP5L
|
SH3 binding domain protein 5 like
|
| 56907
|
SPIRE1
|
spire type actin nucleation factor 1
|
| 8987
|
STBD1
|
starch binding domain 1
|
| 11075
|
STMN2
|
stathmin 2
|
| 171024
|
SYNPO2
|
synaptopodin 2
|
| 25851
|
TECPR1
|
tectonin beta-propeller repeat containing 1
|
| 391123
|
VSIG8
|
V-set and immunoglobulin domain containing 8
|
| 124220
|
ZG16B
|
zymogen granule protein 16B
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
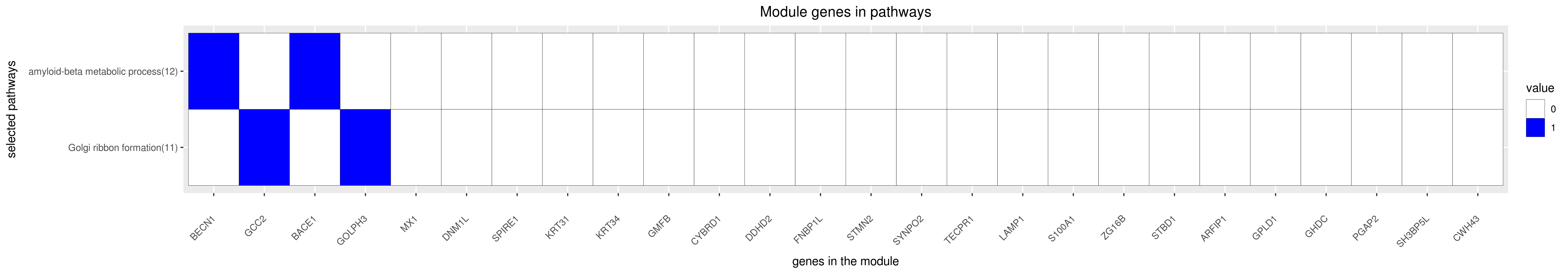
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 4.89e-03
|
4.03e-01
|
ANTIVIRAL MECHANISM BY IFN STIMULATED GENES
|
| 7.53e-03
|
6.38e-01
|
IRON UPTAKE AND TRANSPORT
|
| 1.50e-02
|
1.00e+00
|
ANTIVIRAL MECHANISM BY IFN STIMULATED GENES
|
| 1.70e-02
|
1.00e+00
|
APOPTOTIC EXECUTION PHASE
|
| 1.85e-02
|
1.00e+00
|
IRON UPTAKE AND TRANSPORT
|
| 2.53e-02
|
1.00e+00
|
APOPTOTIC EXECUTION PHASE
|
| 2.77e-02
|
1.00e+00
|
APOPTOSIS
|
| 2.88e-02
|
1.00e+00
|
INTERFERON ALPHA BETA SIGNALING
|
| 3.09e-02
|
1.00e+00
|
APOPTOSIS
|
| 3.45e-02
|
1.00e+00
|
INTERFERON ALPHA BETA SIGNALING
|
| 3.89e-02
|
1.00e+00
|
INTERFERON SIGNALING
|
| 4.41e-02
|
1.00e+00
|
CYTOKINE SIGNALING IN IMMUNE SYSTEM
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:38:10 2018 - R2HTML