Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod371
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod371 |
| Module size |
47 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 10863
|
ADAM23
|
ADAM metallopeptidase domain 28
|
| 10551
|
AGR2
|
anterior gradient 2, protein disulphide isomerase family member
|
| 57463
|
AMIGO1
|
adhesion molecule with Ig like domain 1
|
| 347902
|
AMIGO2
|
adhesion molecule with Ig like domain 2
|
| 386724
|
AMIGO3
|
adhesion molecule with Ig like domain 3
|
| 339201
|
ASB16-AS1
|
ASB16 antisense RNA 1
|
| 8938
|
BAIAP3
|
BAI1 associated protein 3
|
| 8419
|
BFSP2
|
beaded filament structural protein 2
|
| 819
|
CAMLG
|
calcium modulating ligand
|
| 912
|
CD1D
|
CD1d molecule
|
| 976
|
CD97
|
adhesion G protein-coupled receptor E5
|
| 84518
|
CNFN
|
cornifelin
|
| 8292
|
COLQ
|
collagen like tail subunit of asymmetric acetylcholinesterase
|
| 102724652
|
CRYAA
|
crystallin alpha A2
|
| 1411
|
CRYBA1
|
crystallin beta A1
|
| 1412
|
CRYBA2
|
crystallin beta A2
|
| 1417
|
CRYBB3
|
crystallin beta B3
|
| 1420
|
CRYGC
|
crystallin gamma C
|
| 1485
|
CTAG1B
|
cancer/testis antigen 1B
|
| 1509
|
CTSD
|
cathepsin D
|
| 30817
|
EMR2
|
adhesion G protein-coupled receptor E2
|
| 54845
|
ESRP1
|
epithelial splicing regulatory protein 1
|
| 80004
|
ESRP2
|
epithelial splicing regulatory protein 2
|
| 2125
|
EVPL
|
envoplakin
|
| 2350
|
FOLR2
|
folate receptor beta
|
| 27190
|
IL17B
|
interleukin 17B
|
| 79070
|
KDELC1
|
KDEL motif containing 1
|
| 387264
|
KRTAP5-1
|
keratin associated protein 5-1
|
| 10314
|
LANCL1
|
LanC like 1
|
| 10609
|
LEPREL4
|
prolyl 3-hydroxylase family member 4 (non-enzymatic)
|
| 4014
|
LOR
|
loricrin
|
| 26020
|
LRP10
|
LDL receptor related protein 10
|
| 4241
|
MFI2
|
melanotransferrin
|
| 23154
|
NCDN
|
neurochondrin
|
| 9796
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein
|
| 5304
|
PIP
|
prolactin induced protein
|
| 51535
|
PPHLN1
|
periphilin 1
|
| 79170
|
PRR15L
|
proline rich 15 like
|
| 5660
|
PSAP
|
prosaposin
|
| 9783
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
| 138065
|
RNF183
|
ring finger protein 183
|
| 8796
|
SCEL
|
sciellin
|
| 6440
|
SFTPC
|
surfactant protein C
|
| 9368
|
SLC9A3R1
|
SLC9A3 regulator 1
|
| 6699
|
SPRR1B
|
small proline rich protein 1B
|
| 6707
|
SPRR3
|
small proline rich protein 3
|
| 326340
|
ZAR1
|
zygote arrest 1
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
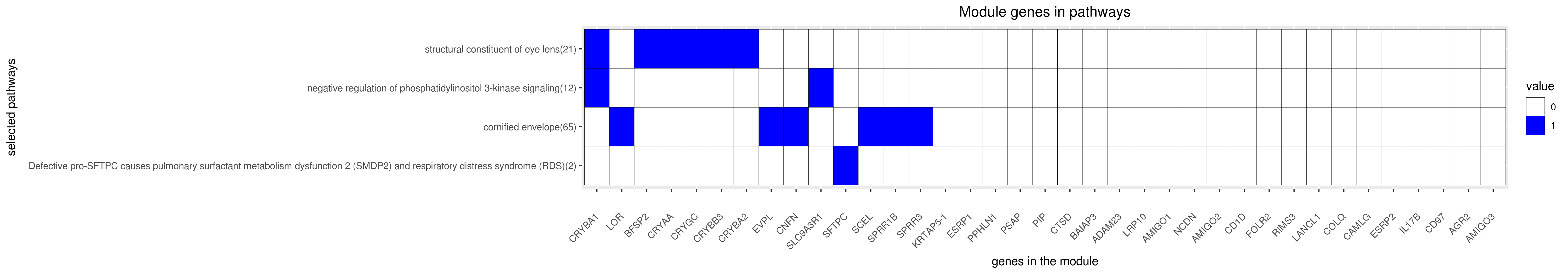
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 1.46e-03
|
1.63e-01
|
CLASS B 2 SECRETIN FAMILY RECEPTORS
|
| 1.10e-02
|
7.90e-01
|
CLASS B 2 SECRETIN FAMILY RECEPTORS
|
| 1.28e-02
|
8.97e-01
|
RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2
|
| 2.33e-02
|
1.00e+00
|
GLYCOSPHINGOLIPID METABOLISM
|
| 2.53e-02
|
1.00e+00
|
SPHINGOLIPID METABOLISM
|
| 3.53e-02
|
1.00e+00
|
GLYCOSPHINGOLIPID METABOLISM
|
| 3.85e-02
|
1.00e+00
|
PLATELET ACTIVATION SIGNALING AND AGGREGATION
|
| 4.42e-02
|
1.00e+00
|
GPCR LIGAND BINDING
|
| 4.71e-02
|
1.00e+00
|
RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:37:56 2018 - R2HTML