Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod366
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod366 |
| Module size |
63 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 389549
|
FEZF1
|
FEZ family zinc finger 1
|
| 55079
|
FEZF2
|
FEZ family zinc finger 2
|
| 2738
|
GLI4
|
GLI family zinc finger 4
|
| 148979
|
GLIS1
|
GLIS family zinc finger 1
|
| 169792
|
GLIS3
|
GLIS family zinc finger 3
|
| 64412
|
GZF1
|
GDNF inducible zinc finger protein 1
|
| 23119
|
HIC2
|
HIC ZBTB transcriptional repressor 2
|
| 3097
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
| 59269
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
| 28999
|
KLF15
|
Kruppel like factor 15
|
| 1316
|
KLF6
|
Kruppel like factor 6
|
| 8609
|
KLF7
|
Kruppel like factor 7
|
| 4150
|
MAZ
|
MYC associated zinc finger protein
|
| 4241
|
MTF1
|
melanotransferrin
|
| 55892
|
MYNN
|
myoneurin
|
| 55922
|
NKRF
|
NFKB repressing factor
|
| 116039
|
OSR2
|
odd-skipped related transciption factor 2
|
| 5325
|
PLAGL1
|
PLAG1 like zinc finger 1
|
| 5326
|
PLAGL2
|
PLAG1 like zinc finger 2
|
| 93166
|
PRDM6
|
PR/SET domain 6
|
| 56978
|
PRDM8
|
PR/SET domain 8
|
| 6239
|
RREB1
|
ras responsive element binding protein 1
|
| 389058
|
SP5
|
Sp5 transcription factor
|
| 121340
|
SP7
|
Sp7 transcription factor
|
| 221833
|
SP8
|
Sp8 transcription factor
|
| 100131390
|
SP9
|
Sp9 transcription factor
|
| 9841
|
ZBTB24
|
zinc finger and BTB domain containing 24
|
| 57684
|
ZBTB26
|
zinc finger and BTB domain containing 26
|
| 79842
|
ZBTB3
|
zinc finger and BTB domain containing 3
|
| 253461
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
| 57659
|
ZBTB4
|
zinc finger and BTB domain containing 4
|
| 23099
|
ZBTB43
|
zinc finger and BTB domain containing 43
|
| 9925
|
ZBTB5
|
zinc finger and BTB domain containing 5
|
| 10773
|
ZBTB6
|
zinc finger and BTB domain containing 6
|
| 653121
|
ZBTB8A
|
zinc finger and BTB domain containing 8A
|
| 132625
|
ZFP42
|
ZFP42 zinc finger protein
|
| 284406
|
ZFP82
|
ZFP82 zinc finger protein
|
| 80829
|
ZFP91
|
ZFP91 zinc finger protein
|
| 84107
|
ZIC4
|
Zic family member 4
|
| 85416
|
ZIC5
|
Zic family member 5
|
| 7694
|
ZNF135
|
zinc finger protein 135
|
| 7707
|
ZNF148
|
zinc finger protein 148
|
| 7757
|
ZNF208
|
zinc finger protein 208
|
| 51222
|
ZNF219
|
zinc finger protein 219
|
| 10793
|
ZNF273
|
zinc finger protein 273
|
| 23528
|
ZNF281
|
zinc finger protein 281
|
| 63925
|
ZNF335
|
zinc finger protein 335
|
| 6940
|
ZNF354A
|
zinc finger protein 354A
|
| 10224
|
ZNF443
|
zinc finger protein 443
|
| 57573
|
ZNF471
|
zinc finger protein 471
|
| 57474
|
ZNF490
|
zinc finger protein 490
|
| 57473
|
ZNF512B
|
zinc finger protein 512B
|
| 130557
|
ZNF513
|
zinc finger protein 513
|
| 162655
|
ZNF519
|
zinc finger protein 519
|
| 51385
|
ZNF589
|
zinc finger protein 589
|
| 22834
|
ZNF652
|
zinc finger protein 652
|
| 65988
|
ZNF747
|
zinc finger protein 747
|
| 92595
|
ZNF764
|
zinc finger protein 764
|
| 374928
|
ZNF773
|
zinc finger protein 773
|
| 54753
|
ZNF853
|
zinc finger protein 853
|
| 7644
|
ZNF91
|
zinc finger protein 91
|
| 7789
|
ZXDA
|
zinc finger X-linked duplicated A
|
| 79364
|
ZXDC
|
ZXD family zinc finger C
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
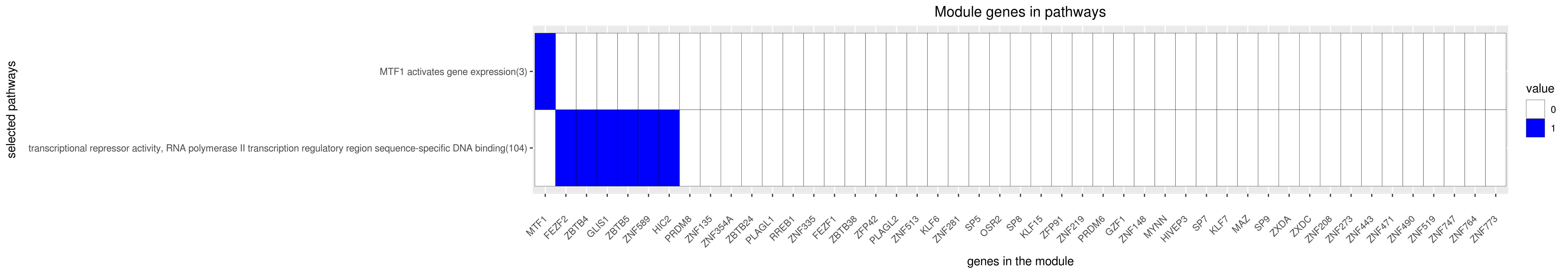
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 2.52e-07
|
6.19e-05
|
GENERIC TRANSCRIPTION PATHWAY
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:37:43 2018 - R2HTML