Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod354
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod354 |
| Module size |
67 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 1961
|
EGR4
|
early growth response 4
|
| 84662
|
GLIS2
|
GLIS family zinc finger 2
|
| 11278
|
KLF12
|
Kruppel like factor 12
|
| 80320
|
KLF14
|
Sp6 transcription factor
|
| 63976
|
PRDM16
|
PR/SET domain 16
|
| 3093
|
UBE2K
|
ubiquitin conjugating enzyme E2 K
|
| 26137
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
| 9880
|
ZBTB39
|
zinc finger and BTB domain containing 39
|
| 9923
|
ZBTB40
|
zinc finger and BTB domain containing 40
|
| 360023
|
ZBTB41
|
zinc finger and BTB domain containing 41
|
| 29068
|
ZBTB44
|
zinc finger and BTB domain containing 44
|
| 221504
|
ZBTB9
|
zinc finger and BTB domain containing 9
|
| 57623
|
ZFAT
|
zinc finger and AT-hook domain containing
|
| 64397
|
ZFP106
|
zinc finger protein 106
|
| 22835
|
ZFP30
|
ZFP30 zinc finger protein
|
| 7539
|
ZFP37
|
ZFP37 zinc finger protein
|
| 146198
|
ZFP90
|
ZFP90 zinc finger protein
|
| 7556
|
ZNF10
|
zinc finger protein 10
|
| 7559
|
ZNF12
|
zinc finger protein 12
|
| 7733
|
ZNF180
|
zinc finger protein 180
|
| 7748
|
ZNF195
|
zinc finger protein 195
|
| 7755
|
ZNF205
|
zinc finger protein 205
|
| 7570
|
ZNF22
|
zinc finger protein 22
|
| 8187
|
ZNF239
|
zinc finger protein 239
|
| 113835
|
ZNF257
|
zinc finger protein 257
|
| 91975
|
ZNF300
|
zinc finger protein 300
|
| 55900
|
ZNF302
|
zinc finger protein 302
|
| 57693
|
ZNF317
|
zinc finger protein 317
|
| 64288
|
ZNF323
|
zinc finger and SCAN domain containing 31
|
| 55713
|
ZNF334
|
zinc finger protein 334
|
| 59348
|
ZNF350
|
zinc finger protein 350
|
| 167465
|
ZNF366
|
zinc finger protein 366
|
| 346157
|
ZNF391
|
zinc finger protein 391
|
| 79744
|
ZNF419
|
zinc finger protein 419
|
| 80818
|
ZNF436
|
zinc finger protein 436
|
| 79973
|
ZNF442
|
zinc finger protein 442
|
| 58499
|
ZNF462
|
zinc finger protein 462
|
| 170958
|
ZNF525
|
zinc finger protein 525
|
| 162972
|
ZNF550
|
zinc finger protein 550
|
| 163081
|
ZNF567
|
zinc finger protein 567
|
| 79177
|
ZNF576
|
zinc finger protein 576
|
| 201514
|
ZNF584
|
zinc finger protein 584
|
| 89887
|
ZNF628
|
zinc finger protein 628
|
| 84146
|
ZNF644
|
zinc finger protein 644
|
| 65251
|
ZNF649
|
zinc finger protein 649
|
| 144348
|
ZNF664
|
zinc finger protein 664
|
| 93474
|
ZNF670
|
zinc finger protein 670
|
| 342926
|
ZNF677
|
zinc finger protein 677
|
| 57116
|
ZNF695
|
zinc finger protein 695
|
| 374879
|
ZNF699
|
zinc finger protein 699
|
| 7562
|
ZNF708
|
zinc finger protein 708
|
| 7552
|
ZNF711
|
zinc finger protein 711
|
| 100131827
|
ZNF717
|
zinc finger protein 717
|
| 654254
|
ZNF732
|
zinc finger protein 732
|
| 126208
|
ZNF787
|
zinc finger protein 787
|
| 285989
|
ZNF789
|
zinc finger protein 789
|
| 90576
|
ZNF799
|
zinc finger protein 799
|
| 7554
|
ZNF8
|
zinc finger protein 8
|
| 7634
|
ZNF80
|
zinc finger protein 80
|
| 125893
|
ZNF816
|
zinc finger protein 816
|
| 55552
|
ZNF823
|
zinc finger protein 823
|
| 284391
|
ZNF844
|
zinc finger protein 844
|
| 643641
|
ZNF862
|
zinc finger protein 862
|
| 7643
|
ZNF90
|
zinc finger protein 90
|
| 79149
|
ZSCAN5A
|
zinc finger and SCAN domain containing 5A
|
| 342933
|
ZSCAN5B
|
zinc finger and SCAN domain containing 5B
|
| 649137
|
ZSCAN5C
|
zinc finger and SCAN domain containing 5C
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
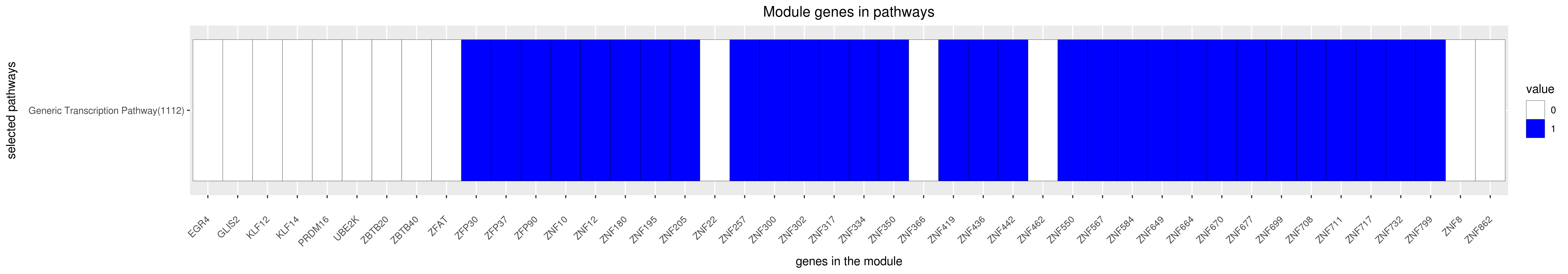
Gene membership
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0.00e+00
|
0.00e+00
|
GENERIC TRANSCRIPTION PATHWAY
|
| 9.02e-03
|
7.37e-01
|
NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING
|
| 1.19e-02
|
9.15e-01
|
RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS
|
| 3.75e-02
|
1.00e+00
|
INNATE IMMUNE SYSTEM
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:37:12 2018 - R2HTML