Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod35
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod35 |
| Module size |
50 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 283847
|
CCDC79
|
telomere repeat binding bouquet formation protein 1
|
| 163126
|
EID2
|
EP300 interacting inhibitor of differentiation 2
|
| 126272
|
EID2B
|
EP300 interacting inhibitor of differentiation 2B
|
| 493861
|
EID3
|
EP300 interacting inhibitor of differentiation 3
|
| 4100
|
MAGEA1
|
MAGE family member A1
|
| 4109
|
MAGEA10
|
MAGE family member A10
|
| 4110
|
MAGEA11
|
MAGE family member A11
|
| 4111
|
MAGEA12
|
MAGE family member A12
|
| 266740
|
MAGEA2
|
MAGE family member A2B
|
| 266740
|
MAGEA2B
|
MAGE family member A2B
|
| 4102
|
MAGEA3
|
MAGE family member A3
|
| 4104
|
MAGEA4
|
MAGE family member A5
|
| 4102
|
MAGEA6
|
MAGE family member A3
|
| 4107
|
MAGEA8
|
MAGE family member A8
|
| 4108
|
MAGEA9
|
MAGE family member A9
|
| 728269
|
MAGEA9B
|
MAGE family member A9B
|
| 4112
|
MAGEB1
|
MAGE family member B1
|
| 139422
|
MAGEB10
|
MAGE family member B10
|
| 139604
|
MAGEB16
|
MAGE family member B16
|
| 645864
|
MAGEB17
|
MAGE family member B17
|
| 286514
|
MAGEB18
|
MAGE family member B18
|
| 4113
|
MAGEB2
|
MAGE family member B2
|
| 4114
|
MAGEB3
|
MAGE family member B3
|
| 4115
|
MAGEB4
|
MAGE family member B4
|
| 347541
|
MAGEB5
|
MAGE family member B5
|
| 158809
|
MAGEB6
|
MAGE family member B6
|
| 9947
|
MAGEC1
|
MAGE family member C1
|
| 51438
|
MAGEC2
|
MAGE family member C2
|
| 139081
|
MAGEC3
|
MAGE family member C3
|
| 9500
|
MAGED1
|
MAGE family member D1
|
| 10916
|
MAGED2
|
MAGE family member D2
|
| 728239
|
MAGED4
|
MAGE family member D4
|
| 81557
|
MAGED4B
|
MAGE family member D4B
|
| 139599
|
MAGEE2
|
MAGE family member E2
|
| 64110
|
MAGEF1
|
MAGE family member F1
|
| 28986
|
MAGEH1
|
MAGE family member H1
|
| 54551
|
MAGEL2
|
MAGE family member L2
|
| 4692
|
NDN
|
necdin, MAGE family member
|
| 56160
|
NDNL2
|
NSE3 homolog, SMC5-SMC6 complex component
|
| 197370
|
NSMCE1
|
NSE1 homolog, SMC5-SMC6 complex component
|
| 286053
|
NSMCE2
|
NSE2 (MMS21) homolog, SMC5-SMC6 complex SUMO ligase
|
| 54780
|
NSMCE4A
|
NSE4 homolog A, SMC5-SMC6 complex component
|
| 22976
|
PAXIP1
|
PAX interacting protein 1
|
| 8243
|
SMC1A
|
structural maintenance of chromosomes 1A
|
| 27127
|
SMC1B
|
structural maintenance of chromosomes 1B
|
| 83452
|
SMC2
|
RAB33B, member RAS oncogene family
|
| 10051
|
SMC4
|
structural maintenance of chromosomes 4
|
| 23137
|
SMC5
|
structural maintenance of chromosomes 5
|
| 79677
|
SMC6
|
structural maintenance of chromosomes 6
|
| 7216
|
TRO
|
trophinin
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
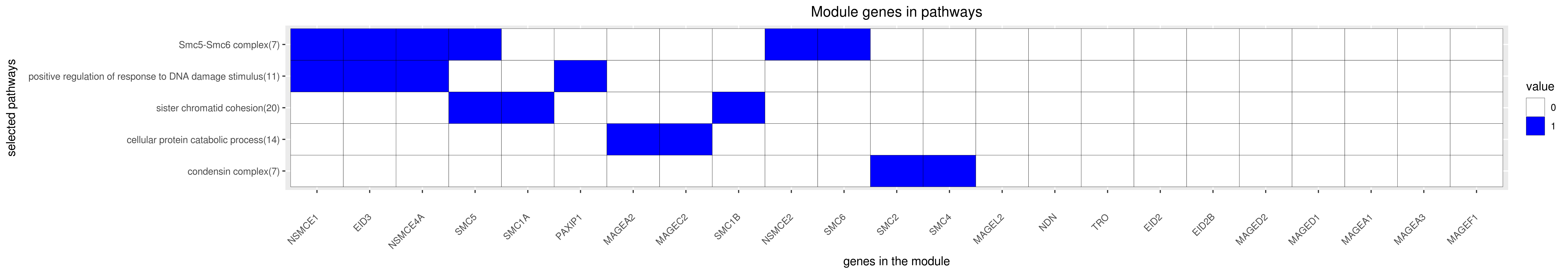
Gene membership
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 2.26e-04
|
2.65e-02
|
MEIOTIC SYNAPSIS
|
| 4.72e-04
|
5.15e-02
|
CHROMOSOME MAINTENANCE
|
| 2.12e-03
|
1.96e-01
|
MEIOSIS
|
| 3.16e-03
|
3.15e-01
|
MEIOTIC SYNAPSIS
|
| 4.68e-03
|
4.33e-01
|
CHROMOSOME MAINTENANCE
|
| 5.15e-03
|
4.69e-01
|
ROLE OF DCC IN REGULATING APOPTOSIS
|
| 5.18e-03
|
4.70e-01
|
REGULATION OF APOPTOSIS
|
| 6.22e-03
|
5.47e-01
|
MEIOSIS
|
| 1.05e-02
|
7.62e-01
|
ROLE OF DCC IN REGULATING APOPTOSIS
|
| 1.14e-02
|
8.16e-01
|
REGULATION OF APOPTOSIS
|
| 2.82e-02
|
1.00e+00
|
CELL CYCLE
|
| 3.91e-02
|
1.00e+00
|
NRAGE SIGNALS DEATH THROUGH JNK
|
| 4.55e-02
|
1.00e+00
|
NRAGE SIGNALS DEATH THROUGH JNK
|
| 4.60e-02
|
1.00e+00
|
APOPTOSIS
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:21:22 2018 - R2HTML