Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod338
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod338 |
| Module size |
87 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 128209
|
KLF17
|
Kruppel like factor 17
|
| 59336
|
PRDM13
|
PR/SET domain 13
|
| 221527
|
ZBTB12
|
zinc finger and BTB domain containing 12
|
| 346171
|
ZFP57
|
ZFP57 zinc finger protein
|
| 386607
|
ZFP91-CNTF
|
ZFP91-CNTF readthrough (NMD candidate)
|
| 114026
|
ZIM3
|
zinc finger imprinted 3
|
| 80317
|
ZKSCAN3
|
zinc finger with KRAB and SCAN domains 3
|
| 7675
|
ZNF121
|
zinc finger protein 121
|
| 7693
|
ZNF134
|
zinc finger protein 134
|
| 7565
|
ZNF17
|
zinc finger protein 17
|
| 339318
|
ZNF181
|
zinc finger protein 181
|
| 7988
|
ZNF182
|
zinc finger protein 212
|
| 7738
|
ZNF184
|
zinc finger protein 184
|
| 105379427
|
ZNF2
|
zinc finger protein 73
|
| 7638
|
ZNF221
|
zinc finger protein 221
|
| 7766
|
ZNF223
|
zinc finger protein 223
|
| 353355
|
ZNF233
|
zinc finger protein 233
|
| 57209
|
ZNF248
|
zinc finger protein 248
|
| 342909
|
ZNF284
|
zinc finger protein 284
|
| 729288
|
ZNF286B
|
zinc finger protein 286B
|
| 90075
|
ZNF30
|
zinc finger protein 30
|
| 282890
|
ZNF311
|
zinc finger protein 311
|
| 162967
|
ZNF320
|
zinc finger protein 320
|
| 399669
|
ZNF321P
|
zinc finger protein 321, pseudogene
|
| 25799
|
ZNF324
|
zinc finger protein 324
|
| 342908
|
ZNF404
|
zinc finger protein 404
|
| 55659
|
ZNF416
|
zinc finger protein 416
|
| 147686
|
ZNF418
|
zinc finger protein 418
|
| 9668
|
ZNF432
|
zinc finger protein 432
|
| 163059
|
ZNF433
|
zinc finger protein 433
|
| 353274
|
ZNF445
|
zinc finger protein 445
|
| 203523
|
ZNF449
|
zinc finger protein 449
|
| 197407
|
ZNF48
|
zinc finger protein 48
|
| 642819
|
ZNF487P
|
zinc finger protein 487
|
| 118738
|
ZNF488
|
zinc finger protein 488
|
| 126069
|
ZNF491
|
zinc finger protein 491
|
| 91392
|
ZNF502
|
zinc finger protein 502
|
| 116115
|
ZNF526
|
zinc finger protein 526
|
| 84503
|
ZNF527
|
zinc finger protein 527
|
| 57711
|
ZNF529
|
zinc finger protein 529
|
| 55205
|
ZNF532
|
zinc finger protein 532
|
| 147658
|
ZNF534
|
zinc finger protein 534
|
| 84215
|
ZNF541
|
zinc finger protein 541
|
| 147694
|
ZNF548
|
zinc finger protein 548
|
| 79230
|
ZNF557
|
zinc finger protein 557
|
| 147837
|
ZNF563
|
zinc finger protein 563
|
| 163050
|
ZNF564
|
zinc finger protein 564
|
| 84924
|
ZNF566
|
zinc finger protein 566
|
| 148268
|
ZNF570
|
zinc finger protein 570
|
| 284346
|
ZNF575
|
zinc finger protein 575
|
| 147949
|
ZNF583
|
zinc finger protein 583
|
| 54807
|
ZNF586
|
zinc finger protein 586
|
| 285267
|
ZNF619
|
zinc finger protein 619
|
| 199692
|
ZNF627
|
zinc finger protein 627
|
| 65243
|
ZNF643
|
ZFP69 zinc finger protein B
|
| 115950
|
ZNF653
|
zinc finger protein 653
|
| 79759
|
ZNF668
|
zinc finger protein 668
|
| 79891
|
ZNF671
|
zinc finger protein 671
|
| 163223
|
ZNF676
|
zinc finger protein 676
|
| 148213
|
ZNF681
|
zinc finger protein 681
|
| 79943
|
ZNF696
|
zinc finger protein 696
|
| 90874
|
ZNF697
|
zinc finger protein 697
|
| 55762
|
ZNF701
|
zinc finger protein 701
|
| 349075
|
ZNF713
|
zinc finger protein 713
|
| 728927
|
ZNF736
|
zinc finger protein 736
|
| 388567
|
ZNF749
|
zinc finger protein 749
|
| 90321
|
ZNF766
|
zinc finger protein 766
|
| 285971
|
ZNF775
|
zinc finger protein 775
|
| 284309
|
ZNF776
|
zinc finger protein 776
|
| 197320
|
ZNF778
|
zinc finger protein 778
|
| 163131
|
ZNF780B
|
zinc finger protein 780B
|
| 163115
|
ZNF781
|
zinc finger protein 781
|
| 158431
|
ZNF782
|
zinc finger protein 782
|
| 147808
|
ZNF784
|
zinc finger protein 784
|
| 388507
|
ZNF788
|
zinc finger family member 788, pseudogene
|
| 168850
|
ZNF800
|
zinc finger protein 800
|
| 374899
|
ZNF829
|
zinc finger protein 829
|
| 128611
|
ZNF831
|
zinc finger protein 831
|
| 90485
|
ZNF835
|
zinc finger protein 835
|
| 116412
|
ZNF837
|
zinc finger protein 837
|
| 285346
|
ZNF852
|
zinc finger protein 852
|
| 729747
|
ZNF878
|
zinc finger protein 878
|
| 400713
|
ZNF880
|
zinc finger protein 880
|
| 101060200
|
ZNF891
|
zinc finger protein 891
|
| 148198
|
ZNF98
|
zinc finger protein 98
|
| 65982
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
| 146050
|
ZSCAN29
|
zinc finger and SCAN domain containing 29
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
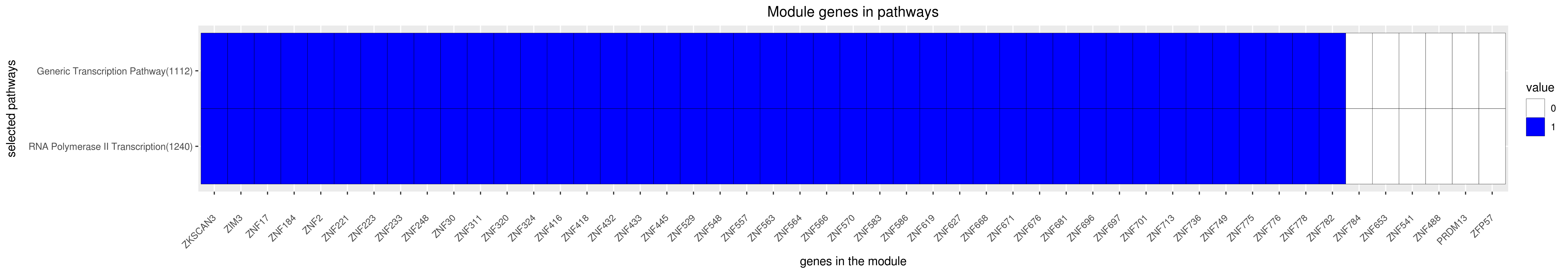
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0e+00
|
0e+00
|
GENERIC TRANSCRIPTION PATHWAY
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:36:23 2018 - R2HTML