Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod337
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod337 |
| Module size |
40 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 203
|
AK1
|
adenylate kinase 1
|
| 204
|
AK2
|
adenylate kinase 2
|
| 50808
|
AK3
|
adenylate kinase 3
|
| 205
|
AK4
|
adenylate kinase 4
|
| 26289
|
AK5
|
adenylate kinase 5
|
| 122481
|
AK7
|
adenylate kinase 7
|
| 158067
|
AK8
|
adenylate kinase 8
|
| 51727
|
CMPK1
|
cytidine/uridine monophosphate kinase 1
|
| 146223
|
CMTM4
|
CKLF like MARVEL transmembrane domain containing 4
|
| 1503
|
CTPS1
|
CTP synthase 1
|
| 56474
|
CTPS2
|
CTP synthase 2
|
| 58511
|
DNASE2B
|
deoxyribonuclease 2 beta
|
| 954
|
ENTPD2
|
ectonucleoside triphosphate diphosphohydrolase 2
|
| 27349
|
MCAT
|
malonyl-CoA-acyl carrier protein transacylase
|
| 4830
|
NME1
|
NME/NM23 nucleoside diphosphate kinase 1
|
| 654364
|
NME1-NME2
|
NME1-NME2 readthrough
|
| 4831
|
NME2
|
NME/NM23 nucleoside diphosphate kinase 2
|
| 4832
|
NME3
|
NME/NM23 nucleoside diphosphate kinase 3
|
| 4833
|
NME4
|
NME/NM23 nucleoside diphosphate kinase 4
|
| 8382
|
NME5
|
NME/NM23 family member 5
|
| 10201
|
NME6
|
NME/NM23 nucleoside diphosphate kinase 6
|
| 29922
|
NME7
|
NME/NM23 family member 7
|
| 347736
|
NME9
|
NME/NM23 family member 9
|
| 84284
|
NTPCR
|
nucleoside-triphosphatase, cancer-related
|
| 87178
|
PNPT1
|
polyribonucleotide nucleotidyltransferase 1
|
| 9533
|
POLR1C
|
RNA polymerase I and III subunit C
|
| 83480
|
PUS3
|
pseudouridylate synthase 3
|
| 126789
|
PUSL1
|
pseudouridylate synthase like 1
|
| 29927
|
SEC61A1
|
Sec61 translocon alpha 1 subunit
|
| 55176
|
SEC61A2
|
Sec61 translocon alpha 2 subunit
|
| 84193
|
SETD3
|
SET domain containing 3
|
| 54093
|
SETD4
|
SET domain containing 4
|
| 6723
|
SRM
|
spermidine synthase
|
| 79188
|
TMEM43
|
transmembrane protein 43
|
| 79799
|
UGT2A3
|
UDP glucuronosyltransferase family 2 member A3
|
| 7365
|
UGT2B10
|
UDP glucuronosyltransferase family 2 member B10
|
| 7366
|
UGT2B15
|
UDP glucuronosyltransferase family 2 member B15
|
| 54490
|
UGT2B28
|
UDP glucuronosyltransferase family 2 member B28
|
| 133688
|
UGT3A1
|
UDP glycosyltransferase family 3 member A1
|
| 167127
|
UGT3A2
|
UDP glycosyltransferase family 3 member A2
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
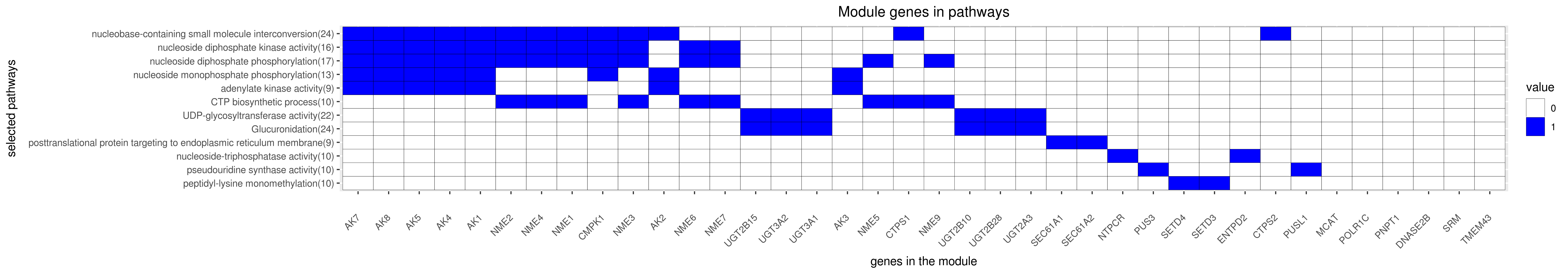
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0.00e+00
|
0.00e+00
|
METABOLISM OF NUCLEOTIDES
|
| 0.00e+00
|
0.00e+00
|
SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES
|
| 0.00e+00
|
0.00e+00
|
METABOLISM OF NUCLEOTIDES
|
| 0.00e+00
|
0.00e+00
|
SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES
|
| 2.57e-04
|
2.98e-02
|
ER PHAGOSOME PATHWAY
|
| 5.12e-04
|
5.54e-02
|
GLUCURONIDATION
|
| 1.08e-03
|
1.26e-01
|
ER PHAGOSOME PATHWAY
|
| 1.14e-03
|
1.13e-01
|
ANTIGEN PROCESSING CROSS PRESENTATION
|
| 4.68e-03
|
4.33e-01
|
ANTIGEN PROCESSING CROSS PRESENTATION
|
| 5.46e-03
|
4.91e-01
|
GLUCURONIDATION
|
| 1.23e-02
|
8.72e-01
|
PHASE II CONJUGATION
|
| 2.12e-02
|
1.00e+00
|
SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE
|
| 2.16e-02
|
1.00e+00
|
RNA POL III CHAIN ELONGATION
|
| 2.16e-02
|
1.00e+00
|
RNA POL III TRANSCRIPTION
|
| 2.16e-02
|
1.00e+00
|
RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER
|
| 2.16e-02
|
1.00e+00
|
RNA POL III TRANSCRIPTION TERMINATION
|
| 2.16e-02
|
1.00e+00
|
RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER
|
| 2.45e-02
|
1.00e+00
|
RNA POL I TRANSCRIPTION TERMINATION
|
| 2.51e-02
|
1.00e+00
|
RNA POL I TRANSCRIPTION TERMINATION
|
| 2.69e-02
|
1.00e+00
|
RNA POL I TRANSCRIPTION INITIATION
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:36:21 2018 - R2HTML