Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod336
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod336 |
| Module size |
30 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 54487
|
DGCR8
|
DGCR8, microprocessor complex subunit
|
| 8446
|
DUSP11
|
dual specificity phosphatase 11
|
| 56339
|
METTL3
|
methyltransferase like 3
|
| 64219
|
PJA1
|
praja ring finger ubiquitin ligase 1
|
| 9867
|
PJA2
|
praja ring finger ubiquitin ligase 2
|
| 5430
|
POLR2A
|
RNA polymerase II subunit A
|
| 5431
|
POLR2B
|
RNA polymerase II subunit B
|
| 5432
|
POLR2C
|
RNA polymerase II subunit C
|
| 51132
|
RLIM
|
ring finger protein, LIM domain interacting
|
| 7844
|
RNF103
|
ring finger protein 103
|
| 79845
|
RNF122
|
ring finger protein 122
|
| 55819
|
RNF130
|
ring finger protein 130
|
| 284996
|
RNF149
|
ring finger protein 149
|
| 57484
|
RNF150
|
ring finger protein 150
|
| 494470
|
RNF165
|
ring finger protein 165
|
| 51255
|
RNF181
|
ring finger protein 181
|
| 200312
|
RNF215
|
ring finger protein 215
|
| 11237
|
RNF24
|
ring finger protein 24
|
| 140545
|
RNF32
|
ring finger protein 32
|
| 152006
|
RNF38
|
ring finger protein 38
|
| 54894
|
RNF43
|
ring finger protein 43
|
| 22838
|
RNF44
|
ring finger protein 44
|
| 6049
|
RNF6
|
ring finger protein 6
|
| 8732
|
RNGTT
|
RNA guanylyltransferase and 5'-phosphatase
|
| 8731
|
RNMT
|
RNA guanine-7 methyltransferase
|
| 79871
|
RPAP2
|
RNA polymerase II associated protein 2
|
| 84138
|
SLC7A6OS
|
solute carrier family 7 member 6 opposite strand
|
| 8725
|
URI1
|
URI1, prefoldin like chaperone
|
| 84133
|
ZNRF3
|
zinc and ring finger 3
|
| 151112
|
ZSWIM2
|
zinc finger SWIM-type containing 2
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
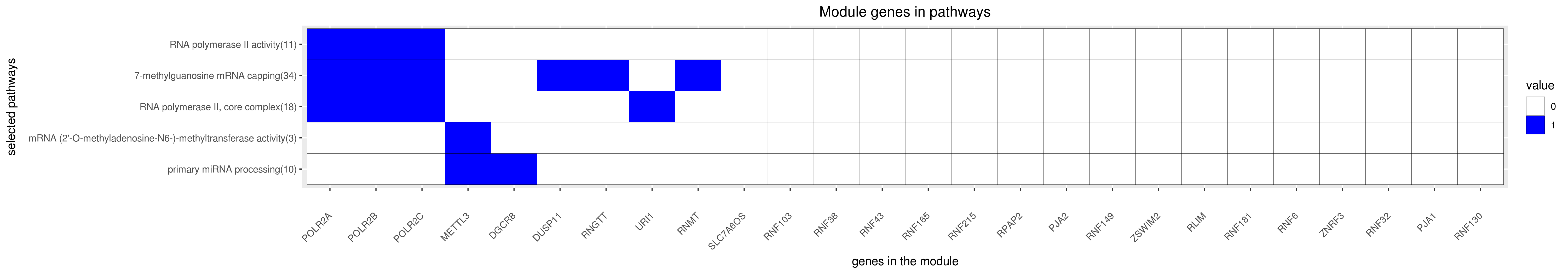
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 8.23e-08
|
2.14e-05
|
MRNA CAPPING
|
| 1.12e-07
|
2.17e-05
|
MRNA CAPPING
|
| 2.22e-07
|
5.50e-05
|
MRNA PROCESSING
|
| 4.22e-07
|
1.00e-04
|
MICRORNA MIRNA BIOGENESIS
|
| 4.38e-07
|
1.04e-04
|
REGULATORY RNA PATHWAYS
|
| 6.13e-07
|
1.42e-04
|
RNA POL II TRANSCRIPTION
|
| 1.06e-06
|
1.83e-04
|
MICRORNA MIRNA BIOGENESIS
|
| 1.10e-06
|
1.89e-04
|
REGULATORY RNA PATHWAYS
|
| 1.41e-06
|
2.39e-04
|
MRNA PROCESSING
|
| 1.45e-06
|
3.19e-04
|
TRANSCRIPTION
|
| 7.65e-06
|
1.49e-03
|
LATE PHASE OF HIV LIFE CYCLE
|
| 1.14e-05
|
2.16e-03
|
HIV LIFE CYCLE
|
| 2.53e-05
|
3.56e-03
|
RNA POL II TRANSCRIPTION
|
| 2.63e-05
|
3.68e-03
|
LATE PHASE OF HIV LIFE CYCLE
|
| 3.53e-05
|
6.11e-03
|
VIRAL MESSENGER RNA SYNTHESIS
|
| 3.99e-05
|
5.44e-03
|
VIRAL MESSENGER RNA SYNTHESIS
|
| 4.03e-05
|
5.48e-03
|
HIV LIFE CYCLE
|
| 5.75e-05
|
9.47e-03
|
ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT
|
| 7.14e-05
|
1.15e-02
|
ELONGATION ARREST AND RECOVERY
|
| 7.77e-05
|
1.24e-02
|
MRNA SPLICING MINOR PATHWAY
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:36:17 2018 - R2HTML