Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod324
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod324 |
| Module size |
39 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 10973
|
ASCC3
|
activating signal cointegrator 1 complex subunit 3
|
| 295334
|
BCAS2
|
BCAS2, pre-mRNA processing factor
|
| 8896
|
BUD31
|
BUD31 homolog
|
| 83938
|
C10orf11
|
leucine rich melanocyte differentiation associated
|
| 151903
|
CCDC12
|
coiled-coil domain containing 12
|
| 55702
|
CCDC94
|
YJU2 splicing factor homolog
|
| 988
|
CDC5L
|
cell division cycle 5 like
|
| 51340
|
CRNKL1
|
crooked neck pre-mRNA splicing factor 1
|
| 57703
|
CWC22
|
CWC22 spliceosome associated protein homolog
|
| 360613
|
CWC25
|
CWC25 spliceosome-associated protein homolog
|
| 1665
|
DHX15
|
DEAH-box helicase 15
|
| 8449
|
DHX16
|
DEAH-box helicase 16
|
| 54505
|
DHX29
|
DExH-box helicase 29
|
| 22907
|
DHX30
|
DExH-box helicase 30
|
| 55760
|
DHX32
|
DEAH-box helicase 32 (putative)
|
| 56919
|
DHX33
|
DEAH-box helicase 33
|
| 9704
|
DHX34
|
DExH-box helicase 34
|
| 60625
|
DHX35
|
DEAH-box helicase 35
|
| 170506
|
DHX36
|
DEAH-box helicase 36
|
| 79665
|
DHX40
|
DEAH-box helicase 40
|
| 90957
|
DHX57
|
DExH-box helicase 57
|
| 1659
|
DHX8
|
DEAH-box helicase 8
|
| 165545
|
DQX1
|
DEAQ-box RNA dependent ATPase 1
|
| 113510
|
HELQ
|
helicase, POLQ like
|
| 164045
|
HFM1
|
HFM1, ATP dependent DNA helicase homolog
|
| 57461
|
ISY1
|
ISY1 splicing factor homolog
|
| 100534599
|
ISY1-RAB43
|
ISY1-RAB43 readthrough
|
| 5356
|
PLRG1
|
pleiotropic regulator 1
|
| 8559
|
PRPF18
|
pre-mRNA processing factor 18
|
| 27339
|
PRPF19
|
pre-mRNA processing factor 19
|
| 55696
|
RBM22
|
RNA binding motif protein 22
|
| 163859
|
SDE2
|
SDE2 telomere maintenance homolog
|
| 22938
|
SNW1
|
SNW domain containing 1
|
| 222183
|
SRRM3
|
serine/arginine repetitive matrix 3
|
| 170933
|
SYF2
|
SYF2 pre-mRNA-splicing factor
|
| 24144
|
TFIP11
|
tuftelin interacting protein 11
|
| 56949
|
XAB2
|
XPA binding protein 2
|
| 64848
|
YTHDC2
|
YTH domain containing 2
|
| 51538
|
ZCCHC17
|
zinc finger CCHC-type containing 17
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
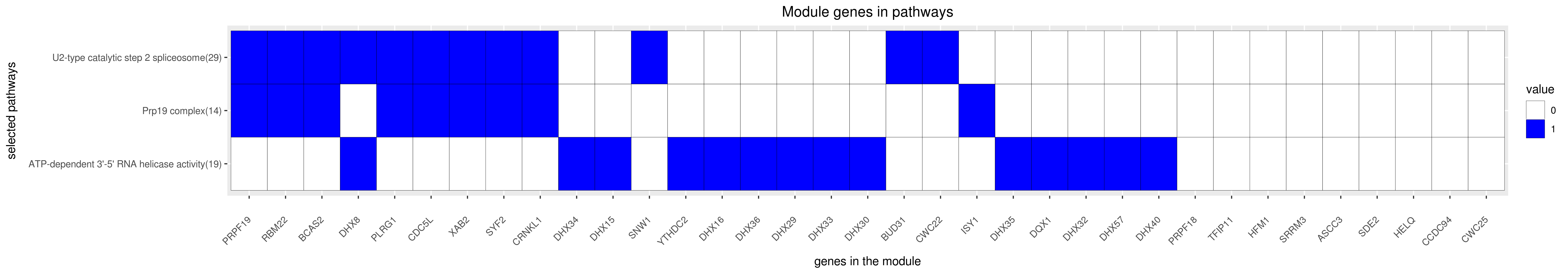
Gene membership
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 3.37e-03
|
3.31e-01
|
FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX
|
| 3.38e-03
|
3.32e-01
|
TRANSCRIPTION COUPLED NER TC NER
|
| 8.33e-03
|
6.93e-01
|
NUCLEOTIDE EXCISION REPAIR
|
| 8.64e-03
|
6.53e-01
|
FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX
|
| 8.67e-03
|
6.55e-01
|
TRANSCRIPTION COUPLED NER TC NER
|
| 9.00e-03
|
6.75e-01
|
NOTCH HLH TRANSCRIPTION PATHWAY
|
| 1.03e-02
|
8.15e-01
|
NOTCH HLH TRANSCRIPTION PATHWAY
|
| 1.42e-02
|
9.73e-01
|
NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION
|
| 1.67e-02
|
1.00e+00
|
NUCLEOTIDE EXCISION REPAIR
|
| 1.68e-02
|
1.00e+00
|
PRE NOTCH TRANSCRIPTION AND TRANSLATION
|
| 1.97e-02
|
1.00e+00
|
SIGNALING BY NOTCH1
|
| 2.02e-02
|
1.00e+00
|
NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION
|
| 2.18e-02
|
1.00e+00
|
PRE NOTCH EXPRESSION AND PROCESSING
|
| 2.26e-02
|
1.00e+00
|
PRE NOTCH TRANSCRIPTION AND TRANSLATION
|
| 2.27e-02
|
1.00e+00
|
PRE NOTCH EXPRESSION AND PROCESSING
|
| 3.03e-02
|
1.00e+00
|
SIGNALING BY NOTCH1
|
| 3.34e-02
|
1.00e+00
|
SIGNALING BY NOTCH
|
| 4.67e-02
|
1.00e+00
|
SIGNALING BY NOTCH
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:35:42 2018 - R2HTML