Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod320
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod320 |
| Module size |
39 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 25999
|
CLIP3
|
CAP-Gly domain containing linker protein 3
|
| 53347
|
CLIP4
|
ubiquitin associated and SH3 domain containing A
|
| 201625
|
DNAH12
|
dynein axonemal heavy chain 12
|
| 154288
|
KHDC3L
|
KH domain containing 3 like, subcortical maternal complex member
|
| 3832
|
KIF11
|
kinesin family member 11
|
| 113220
|
KIF12
|
kinesin family member 12
|
| 63971
|
KIF13A
|
kinesin family member 13A
|
| 23303
|
KIF13B
|
kinesin family member 13B
|
| 9928
|
KIF14
|
kinesin family member 14
|
| 56992
|
KIF15
|
kinesin family member 15
|
| 55614
|
KIF16B
|
kinesin family member 16B
|
| 57576
|
KIF17
|
kinesin family member 17
|
| 124602
|
KIF19
|
kinesin family member 19
|
| 547
|
KIF1A
|
kinesin family member 1A
|
| 305967
|
KIF1B
|
kinesin family member 13B
|
| 10749
|
KIF1C
|
kinesin family member 1C
|
| 9585
|
KIF20B
|
kinesin family member 20B
|
| 55605
|
KIF21A
|
kinesin family member 21A
|
| 23046
|
KIF21B
|
kinesin family member 21B
|
| 3835
|
KIF22
|
kinesin family member 22
|
| 347240
|
KIF24
|
kinesin family member 24
|
| 3834
|
KIF25
|
kinesin family member 25
|
| 55083
|
KIF26B
|
kinesin family member 26B
|
| 55582
|
KIF27
|
kinesin family member 27
|
| 11127
|
KIF3A
|
kinesin family member 3A
|
| 9371
|
KIF3B
|
kinesin family member 3B
|
| 3797
|
KIF3C
|
kinesin family member 3C
|
| 285643
|
KIF4B
|
kinesin family member 4B
|
| 3798
|
KIF5A
|
kinesin family member 5A
|
| 3800
|
KIF5C
|
kinesin family member 5C
|
| 221458
|
KIF6
|
kinesin family member 6
|
| 374654
|
KIF7
|
kinesin family member 7
|
| 64147
|
KIF9
|
kinesin family member 9
|
| 3833
|
KIFC1
|
kinesin family member C1
|
| 90990
|
KIFC2
|
kinesin family member C2
|
| 3801
|
KIFC3
|
kinesin family member C3
|
| 10982
|
MAPRE2
|
microtubule associated protein RP/EB family member 2
|
| 22924
|
MAPRE3
|
microtubule associated protein RP/EB family member 3
|
| 57519
|
STARD9
|
StAR related lipid transfer domain containing 9
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
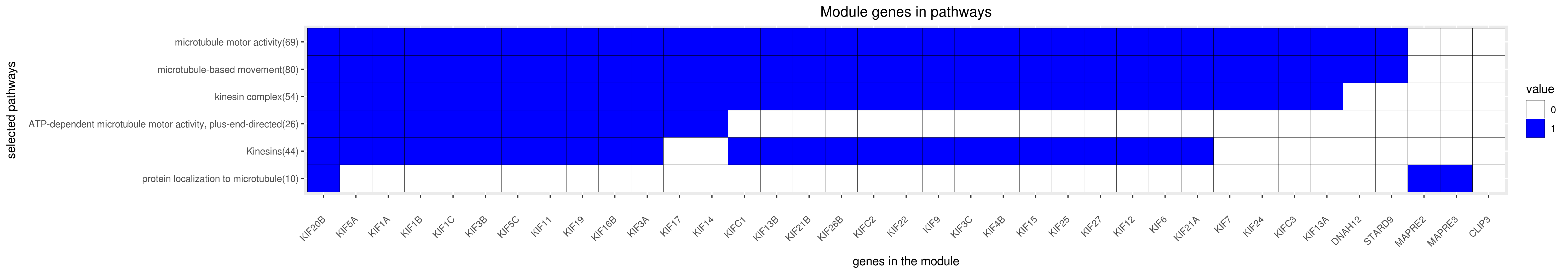
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0.00e+00
|
0.00e+00
|
FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION
|
| 0.00e+00
|
0.00e+00
|
KINESINS
|
| 0.00e+00
|
0.00e+00
|
FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION
|
| 0.00e+00
|
0.00e+00
|
MHC CLASS II ANTIGEN PRESENTATION
|
| 0.00e+00
|
0.00e+00
|
KINESINS
|
| 2.66e-11
|
1.01e-08
|
MHC CLASS II ANTIGEN PRESENTATION
|
| 1.28e-03
|
1.25e-01
|
ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS
|
| 2.18e-03
|
2.30e-01
|
ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS
|
| 4.61e-03
|
3.84e-01
|
PROTEIN FOLDING
|
| 5.05e-03
|
4.61e-01
|
PROTEIN FOLDING
|
| 6.37e-03
|
5.57e-01
|
RECYCLING PATHWAY OF L1
|
| 2.21e-02
|
1.00e+00
|
RECYCLING PATHWAY OF L1
|
| 3.00e-02
|
1.00e+00
|
L1CAM INTERACTIONS
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:35:30 2018 - R2HTML