Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod318
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod318 |
| Module size |
36 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 53947
|
A4GALT
|
alpha 1,4-galactosyltransferase (P blood group)
|
| 2515
|
ADAM2
|
ADAM metallopeptidase domain 2
|
| 8748
|
ADAM20
|
ADAM metallopeptidase domain 20
|
| 8747
|
ADAM21
|
ADAM metallopeptidase domain 21
|
| 11085
|
ADAM30
|
ADAM metallopeptidase domain 30
|
| 175
|
AGA
|
aspartylglucosaminidase
|
| 80150
|
ASRGL1
|
asparaginase like 1
|
| 8706
|
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group)
|
| 2583
|
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyltransferase 1
|
| 9331
|
B4GALT6
|
beta-1,4-galactosyltransferase 6
|
| 1116
|
CHI3L1
|
chitinase 3 like 1
|
| 1117
|
CHI3L2
|
chitinase 3 like 2
|
| 27159
|
CHIA
|
chitinase, acidic
|
| 1118
|
CHIT1
|
chitinase 1
|
| 1476
|
CST6
|
cystatin B
|
| 51076
|
CUTC
|
cutC copper transporter
|
| 64772
|
ENGASE
|
endo-beta-N-acetylglucosaminidase
|
| 2517
|
FUCA1
|
alpha-L-fucosidase 1
|
| 2519
|
FUCA2
|
alpha-L-fucosidase 2
|
| 2530
|
FUT8
|
fucosyltransferase 8
|
| 2720
|
GLB1
|
galactosidase beta 1
|
| 2760
|
GM2A
|
GM2 ganglioside activator
|
| 3073
|
HEXA
|
hexosaminidase subunit alpha
|
| 3074
|
HEXB
|
hexosaminidase subunit beta
|
| 3906
|
LALBA
|
lactalbumin alpha
|
| 3938
|
LCT
|
lactase
|
| 4668
|
NAGA
|
alpha-N-acetylgalactosaminidase
|
| 55577
|
NAGK
|
N-acetylglucosamine kinase
|
| 5016
|
OVGP1
|
oviductal glycoprotein 1
|
| 124912
|
SPACA3
|
sperm acrosome associated 3
|
| 8869
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5
|
| 84302
|
TMEM246
|
transmembrane protein 246
|
| 57829
|
ZP1
|
zona pellucida glycoprotein 4
|
| 7783
|
ZP2
|
zona pellucida glycoprotein 2
|
| 7784
|
ZP3
|
zona pellucida glycoprotein 3
|
| 57829
|
ZP4
|
zona pellucida glycoprotein 4
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
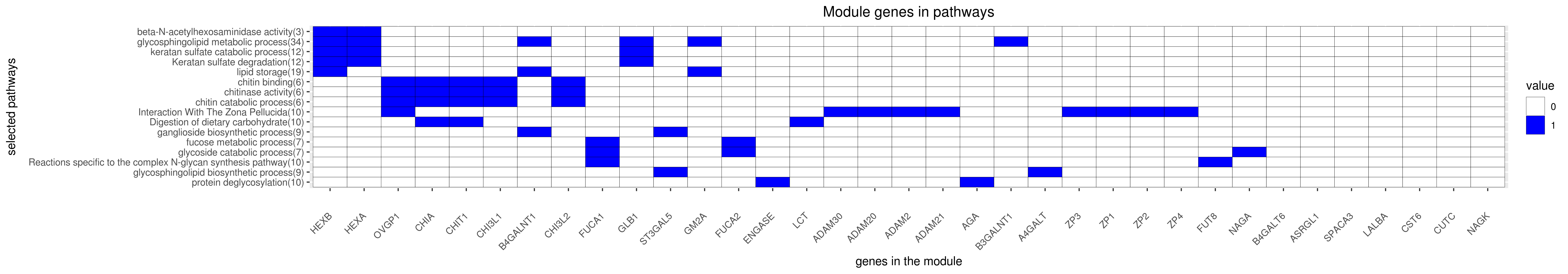
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 1.29e-08
|
2.79e-06
|
GLYCOSPHINGOLIPID METABOLISM
|
| 1.33e-07
|
2.55e-05
|
SPHINGOLIPID METABOLISM
|
| 1.37e-07
|
2.62e-05
|
KERATAN SULFATE KERATIN METABOLISM
|
| 2.29e-07
|
4.29e-05
|
KERATAN SULFATE DEGRADATION
|
| 3.27e-07
|
7.88e-05
|
KERATAN SULFATE KERATIN METABOLISM
|
| 4.78e-07
|
1.13e-04
|
GLYCOSPHINGOLIPID METABOLISM
|
| 1.54e-06
|
3.37e-04
|
KERATAN SULFATE DEGRADATION
|
| 3.32e-06
|
6.85e-04
|
SPHINGOLIPID METABOLISM
|
| 1.17e-05
|
2.21e-03
|
METABOLISM OF CARBOHYDRATES
|
| 1.19e-05
|
1.75e-03
|
METABOLISM OF CARBOHYDRATES
|
| 2.02e-05
|
2.89e-03
|
GLYCOSAMINOGLYCAN METABOLISM
|
| 2.25e-05
|
4.03e-03
|
GLYCOSAMINOGLYCAN METABOLISM
|
| 4.01e-05
|
5.46e-03
|
PHOSPHOLIPID METABOLISM
|
| 1.13e-04
|
1.42e-02
|
HYALURONAN UPTAKE AND DEGRADATION
|
| 1.73e-04
|
2.53e-02
|
PHOSPHOLIPID METABOLISM
|
| 1.80e-04
|
2.16e-02
|
HYALURONAN METABOLISM
|
| 3.70e-04
|
4.93e-02
|
CS DS DEGRADATION
|
| 4.05e-04
|
4.49e-02
|
CS DS DEGRADATION
|
| 4.31e-04
|
5.65e-02
|
N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI
|
| 4.42e-04
|
5.78e-02
|
HYALURONAN UPTAKE AND DEGRADATION
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:35:24 2018 - R2HTML