Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod314
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod314 |
| Module size |
42 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 421
|
ARVCF
|
ARVCF, delta catenin family member
|
| 22926
|
ATF6
|
activating transcription factor 6
|
| 1388
|
ATF6B
|
activating transcription factor 6 beta
|
| 162681
|
C18orf54
|
chromosome 18 open reading frame 54
|
| 55561
|
CDC42BPG
|
CDC42 binding protein kinase gamma
|
| 1496
|
CTNNA2
|
catenin alpha 2
|
| 29119
|
CTNNA3
|
catenin alpha 3
|
| 1501
|
CTNND2
|
catenin delta 2
|
| 51473
|
DCDC2
|
doublecortin domain containing 2
|
| 149069
|
DCDC2B
|
doublecortin domain containing 2B
|
| 728597
|
DCDC2C
|
doublecortin domain containing 2C
|
| 54798
|
DCHS2
|
dachsous cadherin-related 2
|
| 291130
|
DCX
|
doublecortin domain containing 2
|
| 1825
|
DSC1
|
desmocollin 3
|
| 1825
|
DSC2
|
desmocollin 3
|
| 1828
|
DSG1
|
desmoglein 1
|
| 1829
|
DSG2
|
desmoglein 2
|
| 388650
|
FAM69A
|
family with sequence similarity 69 member A
|
| 2196
|
FAT2
|
FAT atypical cadherin 2
|
| 79633
|
FAT4
|
FAT atypical cadherin 4
|
| 2844
|
GPR21
|
G protein-coupled receptor 21
|
| 8994
|
LIMD1
|
LIM domains containing 1
|
| 4026
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
| 92312
|
MEX3A
|
mex-3 RNA binding family member A
|
| 9722
|
NOS1AP
|
nitric oxide synthase 1 adaptor protein
|
| 10038
|
PARP2
|
poly(ADP-ribose) polymerase 2
|
| 10039
|
PARP3
|
poly(ADP-ribose) polymerase family member 3
|
| 5317
|
PKP1
|
plakophilin 1
|
| 5318
|
PKP2
|
plakophilin 2
|
| 11187
|
PKP3
|
plakophilin 3
|
| 8502
|
PKP4
|
plakophilin 4
|
| 5365
|
PLXNB3
|
plexin B3
|
| 266697
|
POM121L4P
|
POM121 transmembrane nucleoporin like 4, pseudogene
|
| 29942
|
PURG
|
purine rich element binding protein G
|
| 94137
|
RP1L1
|
RP1 like 1
|
| 80176
|
SPSB1
|
splA/ryanodine receptor domain and SOCS box containing 1
|
| 92369
|
SPSB4
|
splA/ryanodine receptor domain and SOCS box containing 4
|
| 7205
|
TRIP6
|
thyroid hormone receptor interactor 6
|
| 10497
|
UNC13B
|
unc-13 homolog B
|
| 126374
|
WTIP
|
WT1 interacting protein
|
| 463
|
ZFHX3
|
zinc finger homeobox 3
|
| 79776
|
ZFHX4
|
zinc finger homeobox 4
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
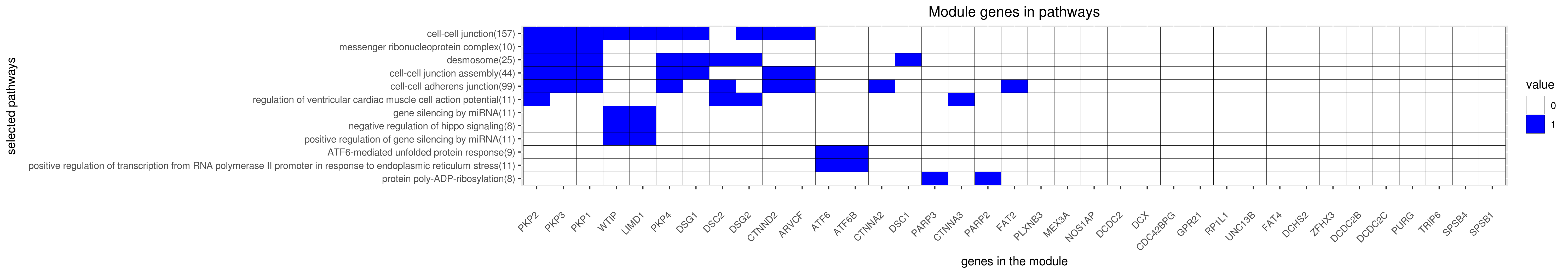
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 1.26e-06
|
2.81e-04
|
APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS
|
| 6.35e-06
|
9.81e-04
|
APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS
|
| 1.87e-05
|
3.40e-03
|
APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS
|
| 4.32e-05
|
7.34e-03
|
APOPTOTIC EXECUTION PHASE
|
| 5.99e-05
|
7.93e-03
|
APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS
|
| 8.68e-05
|
1.12e-02
|
APOPTOTIC EXECUTION PHASE
|
| 1.82e-04
|
2.64e-02
|
APOPTOSIS
|
| 4.66e-04
|
5.09e-02
|
APOPTOSIS
|
| 7.22e-03
|
5.62e-01
|
ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA
|
| 8.89e-03
|
6.68e-01
|
ACTIVATION OF CHAPERONES BY ATF6 ALPHA
|
| 1.00e-02
|
7.32e-01
|
GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE
|
| 1.03e-02
|
8.15e-01
|
ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA
|
| 1.24e-02
|
9.44e-01
|
ACTIVATION OF CHAPERONES BY ATF6 ALPHA
|
| 1.47e-02
|
1.00e+00
|
OTHER SEMAPHORIN INTERACTIONS
|
| 2.36e-02
|
1.00e+00
|
ACTIVATION OF GENES BY ATF4
|
| 2.44e-02
|
1.00e+00
|
PERK REGULATED GENE EXPRESSION
|
| 2.52e-02
|
1.00e+00
|
ACTIVATION OF GENES BY ATF4
|
| 2.68e-02
|
1.00e+00
|
PERK REGULATED GENE EXPRESSION
|
| 2.85e-02
|
1.00e+00
|
ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION
|
| 2.85e-02
|
1.00e+00
|
OTHER SEMAPHORIN INTERACTIONS
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:35:12 2018 - R2HTML