Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod308
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod308 |
| Module size |
41 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 170689
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif 15
|
| 213
|
ALB
|
albumin
|
| 200576
|
CFD
|
phosphoinositide kinase, FYVE-type zinc finger containing
|
| 303300
|
CLU
|
clustered mitochondria homolog
|
| 2162
|
F13A1
|
coagulation factor XIII A chain
|
| 2153
|
F5
|
coagulation factor V
|
| 2277
|
FIGF
|
vascular endothelial growth factor D
|
| 2621
|
GAS6
|
growth arrest specific 6
|
| 3084
|
HRG
|
neuregulin 1
|
| 3479
|
IGF1
|
insulin like growth factor 1
|
| 3481
|
IGF2
|
insulin like growth factor 2
|
| 3484
|
IGFBP1
|
insulin like growth factor binding protein 1
|
| 3486
|
IGFBP3
|
insulin like growth factor binding protein 3
|
| 3487
|
IGFBP4
|
insulin like growth factor binding protein 4
|
| 3488
|
IGFBP5
|
insulin like growth factor binding protein 5
|
| 3489
|
IGFBP6
|
insulin like growth factor binding protein 6
|
| 7044
|
LEFTY2
|
left-right determination factor 2
|
| 4054
|
LTBP3
|
latent transforming growth factor beta binding protein 3
|
| 22915
|
MMRN1
|
multimerin 1
|
| 5069
|
PAPPA
|
pappalysin 1
|
| 60676
|
PAPPA2
|
pappalysin 2
|
| 5196
|
PF4
|
platelet factor 4
|
| 5553
|
PRG2
|
proteoglycan 2, pro eosinophil major basic protein
|
| 5627
|
PROS1
|
protein S
|
| 5265
|
SERPINA1
|
serpin family A member 1
|
| 5054
|
SERPINE1
|
serpin family E member 1
|
| 5345
|
SERPINF2
|
serpin family F member 2
|
| 710
|
SERPING1
|
serpin family G member 1
|
| 6678
|
SPARC
|
secreted protein acidic and cysteine rich
|
| 5552
|
SRGN
|
serglycin
|
| 7042
|
TGFB2
|
transforming growth factor beta 2
|
| 7043
|
TGFB3
|
transforming growth factor beta 3
|
| 7056
|
THBD
|
thrombomodulin
|
| 7076
|
TIMP1
|
TIMP metallopeptidase inhibitor 1
|
| 7114
|
TMSB4X
|
thymosin beta 4 X-linked
|
| 7148
|
TNXB
|
tenascin XB
|
| 7422
|
VEGFA
|
vascular endothelial growth factor A
|
| 7423
|
VEGFB
|
vascular endothelial growth factor B
|
| 7424
|
VEGFC
|
vascular endothelial growth factor C
|
| 7450
|
VWF
|
von Willebrand factor
|
| 8839
|
WISP2
|
WNT1 inducible signaling pathway protein 2
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
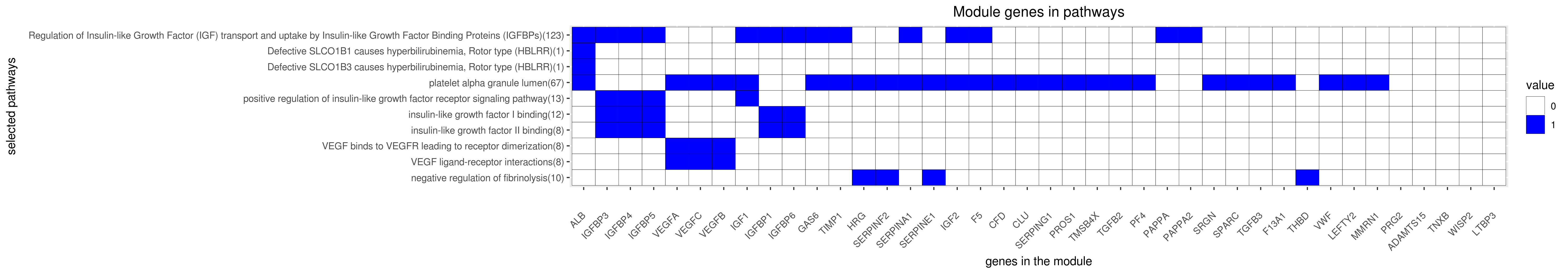
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0.00e+00
|
0.00e+00
|
REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS
|
| 0.00e+00
|
0.00e+00
|
RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2
|
| 0.00e+00
|
0.00e+00
|
PLATELET ACTIVATION SIGNALING AND AGGREGATION
|
| 0.00e+00
|
0.00e+00
|
REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS
|
| 0.00e+00
|
0.00e+00
|
RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2
|
| 0.00e+00
|
0.00e+00
|
PLATELET ACTIVATION SIGNALING AND AGGREGATION
|
| 1.05e-09
|
3.46e-07
|
FORMATION OF FIBRIN CLOT CLOTTING CASCADE
|
| 1.35e-09
|
3.24e-07
|
FORMATION OF FIBRIN CLOT CLOTTING CASCADE
|
| 2.11e-08
|
4.46e-06
|
COMMON PATHWAY
|
| 5.28e-08
|
1.41e-05
|
COMMON PATHWAY
|
| 8.96e-08
|
1.75e-05
|
VEGF LIGAND RECEPTOR INTERACTIONS
|
| 1.26e-06
|
2.15e-04
|
DIABETES PATHWAYS
|
| 1.80e-06
|
3.87e-04
|
VEGF LIGAND RECEPTOR INTERACTIONS
|
| 3.05e-06
|
6.33e-04
|
DIABETES PATHWAYS
|
| 5.14e-03
|
4.68e-01
|
INTRINSIC PATHWAY
|
| 7.42e-03
|
5.74e-01
|
INTRINSIC PATHWAY
|
| 1.05e-02
|
7.62e-01
|
CELL SURFACE INTERACTIONS AT THE VASCULAR WALL
|
| 1.56e-02
|
1.00e+00
|
COMPLEMENT CASCADE
|
| 2.13e-02
|
1.00e+00
|
CELL SURFACE INTERACTIONS AT THE VASCULAR WALL
|
| 2.36e-02
|
1.00e+00
|
COMPLEMENT CASCADE
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:34:55 2018 - R2HTML