Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod289
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod289 |
| Module size |
45 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 113622
|
ADPRHL1
|
ADP-ribosylhydrolase like 1
|
| 57538
|
ALPK3
|
alpha kinase 3
|
| 10930
|
APOBEC2
|
apolipoprotein B mRNA editing enzyme catalytic subunit 2
|
| 800
|
CALD1
|
caldesmon 1
|
| 845
|
CASQ2
|
calsequestrin 2
|
| 286097
|
EFHA2
|
mitochondrial calcium uptake family member 3
|
| 171484
|
FAM9C
|
family with sequence similarity 9 member C
|
| 3270
|
HRC
|
histidine rich calcium binding protein
|
| 91156
|
IGFN1
|
immunoglobulin-like and fibronectin type III domain containing 1
|
| 283284
|
IGSF22
|
immunoglobulin superfamily member 22
|
| 25802
|
LMOD1
|
leiomodin 1
|
| 442721
|
LMOD2
|
leiomodin 2
|
| 343263
|
MYBPHL
|
myosin binding protein H like
|
| 4619
|
MYH1
|
myosin heavy chain 1
|
| 4628
|
MYH10
|
myosin heavy chain 10
|
| 4629
|
MYH11
|
myosin heavy chain 11
|
| 8735
|
MYH13
|
myosin heavy chain 13
|
| 57644
|
MYH14
|
myosin heavy chain 7B
|
| 4620
|
MYH2
|
myosin heavy chain 2
|
| 4625
|
MYH7
|
myosin heavy chain 7
|
| 4627
|
MYH9
|
myosin heavy chain 9
|
| 93408
|
MYL10
|
myosin light chain 10
|
| 10627
|
MYL12A
|
myosin light chain 12A
|
| 103910
|
MYL12B
|
myosin light chain 12B
|
| 4636
|
MYL5
|
myosin light chain 5
|
| 4637
|
MYL6
|
myosin light chain 6
|
| 140465
|
MYL6B
|
myosin light chain 6B
|
| 58498
|
MYL7
|
myosin light chain 7
|
| 10398
|
MYL9
|
myosin light chain 9
|
| 29895
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle
|
| 8736
|
MYOM1
|
myomesin 1
|
| 9499
|
MYOT
|
myotilin
|
| 58529
|
MYOZ1
|
myozenin 1
|
| 23363
|
OBSL1
|
obscurin like 1
|
| 9033
|
PKD2L1
|
polycystin 2 like 1, transient receptor potential cation channel
|
| 134549
|
SHROOM1
|
shroom family member 1
|
| 57477
|
SHROOM4
|
shroom family member 4
|
| 23676
|
SMPX
|
small muscle protein X-linked
|
| 6345
|
SRL
|
sarcalumenin
|
| 29765
|
TMOD4
|
tropomodulin 4
|
| 7134
|
TNNC1
|
troponin C1, slow skeletal and cardiac type
|
| 7125
|
TNNC2
|
troponin C2, fast skeletal type
|
| 10345
|
TRDN
|
triadin
|
| 140803
|
TRPM6
|
transient receptor potential cation channel subfamily M member 6
|
| 80737
|
VWA7
|
von Willebrand factor A domain containing 7
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
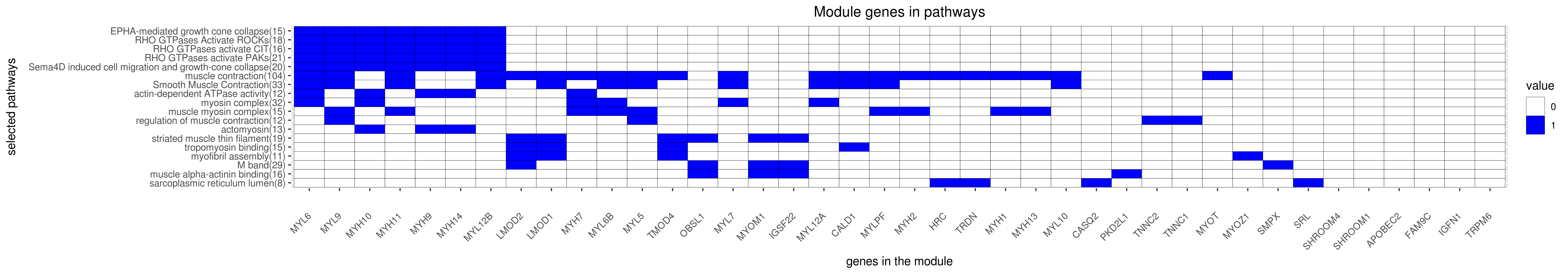
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0.00e+00
|
0.00e+00
|
MUSCLE CONTRACTION
|
| 0.00e+00
|
0.00e+00
|
SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE
|
| 0.00e+00
|
0.00e+00
|
SEMA4D IN SEMAPHORIN SIGNALING
|
| 0.00e+00
|
0.00e+00
|
SMOOTH MUSCLE CONTRACTION
|
| 0.00e+00
|
0.00e+00
|
MUSCLE CONTRACTION
|
| 0.00e+00
|
0.00e+00
|
SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE
|
| 0.00e+00
|
0.00e+00
|
SEMA4D IN SEMAPHORIN SIGNALING
|
| 0.00e+00
|
0.00e+00
|
SMOOTH MUSCLE CONTRACTION
|
| 8.78e-11
|
2.41e-08
|
SEMAPHORIN INTERACTIONS
|
| 2.23e-10
|
7.87e-08
|
SEMAPHORIN INTERACTIONS
|
| 2.22e-07
|
4.17e-05
|
AXON GUIDANCE
|
| 1.64e-06
|
3.56e-04
|
AXON GUIDANCE
|
| 4.58e-05
|
7.71e-03
|
DEVELOPMENTAL BIOLOGY
|
| 4.22e-03
|
3.98e-01
|
STRIATED MUSCLE CONTRACTION
|
| 4.53e-03
|
3.78e-01
|
STRIATED MUSCLE CONTRACTION
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:33:59 2018 - R2HTML