Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod275
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod275 |
| Module size |
31 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 15
|
AANAT
|
aralkylamine N-acetyltransferase
|
| 79370
|
BCL2L14
|
BCL2 like 14
|
| 666
|
BOK
|
BCL2 family apoptosis regulator BOK
|
| 81576
|
CCDC130
|
coiled-coil domain containing 130
|
| 130940
|
CCDC148
|
coiled-coil domain containing 148
|
| 1656
|
DDX6
|
DEAD-box helicase 6
|
| 9988
|
DMTF1
|
cyclin D binding myb like transcription factor 1
|
| 138311
|
FAM69B
|
family with sequence similarity 69 member B
|
| 64841
|
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1
|
| 3831
|
KLC1
|
kinesin light chain 1
|
| 147700
|
KLC2
|
kinesin light chain 3
|
| 147700
|
KLC3
|
kinesin light chain 3
|
| 89953
|
KLC4
|
kinesin light chain 4
|
| 4602
|
MYB
|
MYB proto-oncogene, transcription factor
|
| 4605
|
MYBL2
|
MYB proto-oncogene like 2
|
| 4763
|
NF1
|
neurofibromin 1
|
| 23207
|
PLEKHM2
|
pleckstrin homology and RUN domain containing M2
|
| 51426
|
POLQ
|
DNA polymerase kappa
|
| 5478
|
PPIA
|
peptidylprolyl isomerase A
|
| 653505
|
PPIAL4A
|
peptidylprolyl isomerase A like 4A
|
| 653505
|
PPIAL4B
|
peptidylprolyl isomerase A like 4A
|
| 653598
|
PPIAL4C
|
peptidylprolyl isomerase A like 4C
|
| 645142
|
PPIAL4D
|
peptidylprolyl isomerase A like 4D
|
| 644591
|
PPIAL4G
|
peptidylprolyl isomerase A like 4G
|
| 10450
|
PPIE
|
peptidylprolyl isomerase E
|
| 6992
|
PPP1R11
|
protein phosphatase 1 regulatory inhibitor subunit 11
|
| 23369
|
PUM2
|
pumilio RNA binding family member 2
|
| 10861
|
SAT1
|
solute carrier family 26 member 1
|
| 340562
|
SATL1
|
spermidine/spermine N1-acetyl transferase like 1
|
| 6621
|
SNAPC4
|
small nuclear RNA activating complex polypeptide 4
|
| 55133
|
SRBD1
|
S1 RNA binding domain 1
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
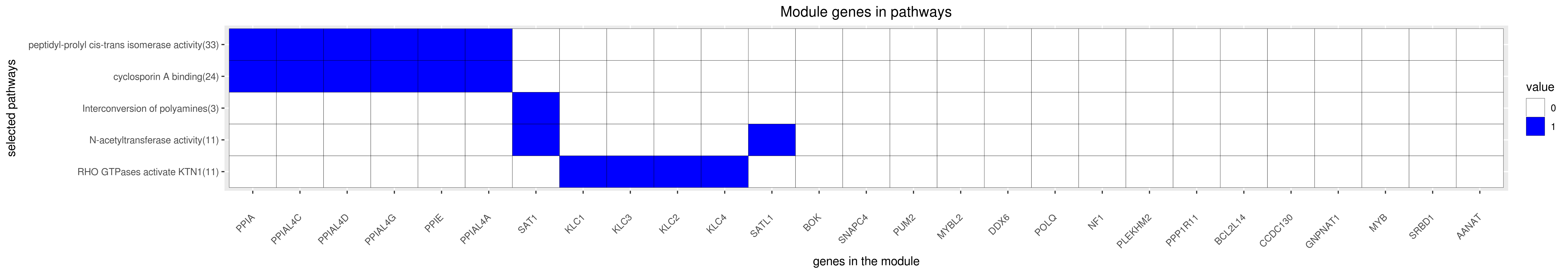
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 1.17e-07
|
2.26e-05
|
KINESINS
|
| 1.93e-07
|
4.84e-05
|
KINESINS
|
| 3.86e-07
|
9.23e-05
|
FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION
|
| 6.18e-07
|
1.11e-04
|
FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION
|
| 5.15e-06
|
1.03e-03
|
MHC CLASS II ANTIGEN PRESENTATION
|
| 1.40e-05
|
2.04e-03
|
MHC CLASS II ANTIGEN PRESENTATION
|
| 1.81e-03
|
1.71e-01
|
INTEGRATION OF PROVIRUS
|
| 3.43e-03
|
3.35e-01
|
BINDING AND ENTRY OF HIV VIRION
|
| 3.62e-03
|
3.12e-01
|
BINDING AND ENTRY OF HIV VIRION
|
| 6.64e-03
|
5.24e-01
|
APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION
|
| 6.86e-03
|
5.92e-01
|
INTEGRATION OF PROVIRUS
|
| 7.05e-03
|
6.04e-01
|
APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION
|
| 7.23e-03
|
5.62e-01
|
EARLY PHASE OF HIV LIFE CYCLE
|
| 8.57e-03
|
7.08e-01
|
EARLY PHASE OF HIV LIFE CYCLE
|
| 1.98e-02
|
1.00e+00
|
BASIGIN INTERACTIONS
|
| 2.75e-02
|
1.00e+00
|
AMINE DERIVED HORMONES
|
| 3.15e-02
|
1.00e+00
|
BASIGIN INTERACTIONS
|
| 3.19e-02
|
1.00e+00
|
RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER
|
| 3.29e-02
|
1.00e+00
|
AMINE DERIVED HORMONES
|
| 3.42e-02
|
1.00e+00
|
HOST INTERACTIONS OF HIV FACTORS
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:33:18 2018 - R2HTML