Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod267
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod267 |
| Module size |
29 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 57698
|
KIAA1598
|
shootin 1
|
| 5954
|
RCN1
|
reticulocalbin 1
|
| 79024
|
SMIM2
|
small integral membrane protein 2
|
| 374768
|
SPEM1
|
spermatid maturation 1
|
| 140809
|
SRXN1
|
sulfiredoxin 1
|
| 10534
|
SSSCA1
|
Sjogren syndrome/scleroderma autoantigen 1
|
| 6817
|
SULT1A1
|
sulfotransferase family 1A member 1
|
| 80213
|
TM2D3
|
TM2 domain containing 3
|
| 55374
|
TMCO6
|
transmembrane and coiled-coil domains 6
|
| 55529
|
TMEM55A
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 2
|
| 90809
|
TMEM55B
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1
|
| 7265
|
TTC1
|
tetratricopeptide repeat domain 1
|
| 283237
|
TTC9C
|
tetratricopeptide repeat domain 9C
|
| 23170
|
TTLL12
|
tubulin tyrosine ligase like 12
|
| 10277
|
UBE4B
|
ubiquitination factor E4B
|
| 84993
|
UBL7
|
ubiquitin like 7
|
| 29979
|
UBQLN1
|
ubiquilin 1
|
| 29978
|
UBQLN2
|
ubiquilin 2
|
| 56893
|
UBQLN4
|
ubiquilin 4
|
| 23304
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2
|
| 23352
|
UBR4
|
ubiquitin protein ligase E3 component n-recognin 4
|
| 51035
|
UBXN1
|
UBX domain protein 1
|
| 7347
|
UCHL3
|
ubiquitin C-terminal hydrolase L3
|
| 445268
|
UFC1
|
ubiquitin-fold modifier conjugating enzyme 1
|
| 51569
|
UFM1
|
ubiquitin fold modifier 1
|
| 85451
|
UNK
|
unkempt family zinc finger
|
| 9736
|
USP34
|
ubiquitin specific peptidase 34
|
| 8239
|
USP9X
|
ubiquitin specific peptidase 9 X-linked
|
| 9406
|
ZRANB2
|
zinc finger RANBP2-type containing 2
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
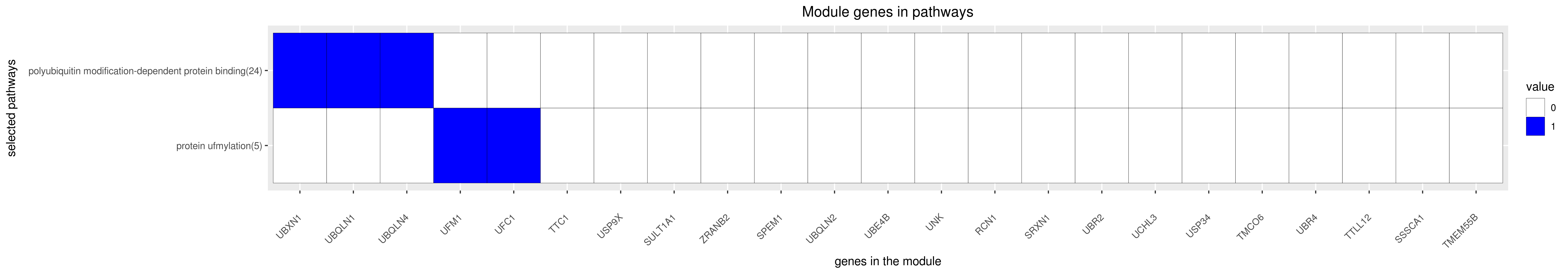
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 3.64e-03
|
3.51e-01
|
RECYCLING PATHWAY OF L1
|
| 7.25e-03
|
6.18e-01
|
DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY
|
| 7.25e-03
|
6.18e-01
|
CYTOSOLIC SULFONATION OF SMALL MOLECULES
|
| 7.53e-03
|
5.81e-01
|
RECYCLING PATHWAY OF L1
|
| 1.00e-02
|
7.32e-01
|
DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY
|
| 1.08e-02
|
8.46e-01
|
ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION
|
| 1.09e-02
|
8.53e-01
|
TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER
|
| 1.12e-02
|
8.71e-01
|
CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION
|
| 1.45e-02
|
1.00e+00
|
SIGNALING BY TGF BETA RECEPTOR COMPLEX
|
| 1.50e-02
|
1.00e+00
|
ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION
|
| 1.57e-02
|
1.00e+00
|
PHASE II CONJUGATION
|
| 1.75e-02
|
1.00e+00
|
TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER
|
| 1.80e-02
|
1.00e+00
|
CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION
|
| 2.72e-02
|
1.00e+00
|
CYTOSOLIC SULFONATION OF SMALL MOLECULES
|
| 3.41e-02
|
1.00e+00
|
SIGNALING BY TGF BETA RECEPTOR COMPLEX
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:32:55 2018 - R2HTML