Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod256
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod256 |
| Module size |
32 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 10815
|
CPLX1
|
complexin 1
|
| 54808
|
DYM
|
dymeclin
|
| 25924
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
| 150221
|
RIMBP3C
|
RIMS binding protein 3C
|
| 22999
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
| 9699
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
| 57030
|
SLC17A7
|
solute carrier family 17 member 7
|
| 6571
|
SLC18A2
|
solute carrier family 18 member A2
|
| 6572
|
SLC18A3
|
solute carrier family 18 member A3
|
| 6616
|
SNAP25
|
synaptosome associated protein 25
|
| 29091
|
STXBP6
|
syntaxin binding protein 6
|
| 9145
|
SYNGR1
|
synaptogyrin 1
|
| 9143
|
SYNGR3
|
synaptogyrin 3
|
| 132204
|
SYNPR
|
synaptoporin
|
| 6855
|
SYP
|
synaptophysin
|
| 6856
|
SYPL1
|
synaptophysin like 1
|
| 6857
|
SYT1
|
synaptotagmin 1
|
| 23208
|
SYT11
|
synaptotagmin 11
|
| 83849
|
SYT15
|
synaptotagmin 15
|
| 51760
|
SYT17
|
synaptotagmin 17
|
| 127833
|
SYT2
|
synaptotagmin 2
|
| 84258
|
SYT3
|
synaptotagmin 3
|
| 6860
|
SYT4
|
synaptotagmin 4
|
| 6861
|
SYT5
|
synaptotagmin 5
|
| 9066
|
SYT7
|
synaptotagmin 7
|
| 90019
|
SYT8
|
synaptotagmin 8
|
| 84958
|
SYTL1
|
synaptotagmin like 1
|
| 54843
|
SYTL2
|
synaptotagmin like 2
|
| 94120
|
SYTL3
|
synaptotagmin like 3
|
| 94121
|
SYTL4
|
synaptotagmin like 4
|
| 94122
|
SYTL5
|
synaptotagmin like 5
|
| 123036
|
TC2N
|
tandem C2 domains, nuclear
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
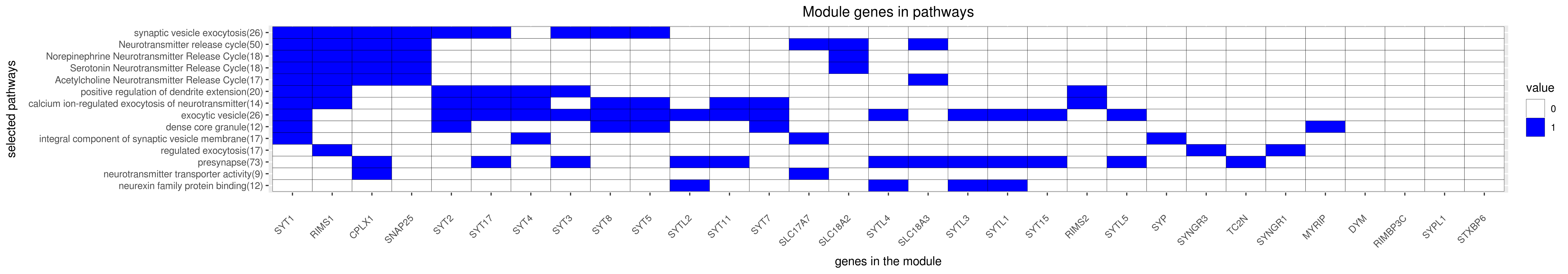
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 2.58e-11
|
9.79e-09
|
NEUROTRANSMITTER RELEASE CYCLE
|
| 4.43e-10
|
1.13e-07
|
NEUROTRANSMITTER RELEASE CYCLE
|
| 1.16e-09
|
3.80e-07
|
NEURONAL SYSTEM
|
| 2.40e-09
|
7.51e-07
|
DOPAMINE NEUROTRANSMITTER RELEASE CYCLE
|
| 3.46e-09
|
1.06e-06
|
ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE
|
| 3.75e-09
|
1.14e-06
|
NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE
|
| 3.39e-08
|
7.03e-06
|
DOPAMINE NEUROTRANSMITTER RELEASE CYCLE
|
| 3.50e-08
|
9.56e-06
|
GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE
|
| 4.07e-08
|
1.10e-05
|
TRANSMISSION ACROSS CHEMICAL SYNAPSES
|
| 4.76e-08
|
9.68e-06
|
NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE
|
| 5.41e-08
|
1.09e-05
|
ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE
|
| 7.54e-08
|
1.49e-05
|
NEURONAL SYSTEM
|
| 2.86e-07
|
5.33e-05
|
GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE
|
| 9.39e-07
|
1.63e-04
|
BOTULINUM NEUROTOXICITY
|
| 1.67e-06
|
3.61e-04
|
BOTULINUM NEUROTOXICITY
|
| 2.35e-06
|
3.85e-04
|
TRANSMISSION ACROSS CHEMICAL SYNAPSES
|
| 6.76e-06
|
1.33e-03
|
GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION
|
| 2.71e-05
|
3.79e-03
|
GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION
|
| 3.99e-03
|
3.79e-01
|
REGULATION OF INSULIN SECRETION
|
| 6.60e-03
|
5.73e-01
|
INTEGRATION OF ENERGY METABOLISM
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:32:23 2018 - R2HTML