Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod25
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod25 |
| Module size |
63 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 23529
|
CLCF1
|
cardiotrophin like cytokine factor 1
|
| 1271
|
CNTFR
|
ciliary neurotrophic factor receptor
|
| 64109
|
CRLF2
|
cytokine receptor like factor 2
|
| 1440
|
CSF3
|
colony stimulating factor 3
|
| 1441
|
CSF3R
|
colony stimulating factor 3 receptor
|
| 1442
|
CSH1
|
chorionic somatomammotropin hormone 1
|
| 1443
|
CSH2
|
chorionic somatomammotropin hormone 2
|
| 1489
|
CTF1
|
cardiotrophin 1
|
| 2689
|
GH2
|
growth hormone 2
|
| 3439
|
IFNA1
|
interferon alpha 1
|
| 3446
|
IFNA10
|
interferon alpha 10
|
| 3447
|
IFNA13
|
interferon alpha 13
|
| 3448
|
IFNA14
|
interferon alpha 14
|
| 3449
|
IFNA16
|
interferon alpha 16
|
| 3451
|
IFNA17
|
interferon alpha 17
|
| 3440
|
IFNA2
|
interferon alpha 2
|
| 3452
|
IFNA21
|
interferon alpha 21
|
| 3441
|
IFNA4
|
interferon alpha 4
|
| 3442
|
IFNA5
|
interferon alpha 5
|
| 3443
|
IFNA6
|
interferon alpha 6
|
| 3444
|
IFNA7
|
interferon alpha 7
|
| 3445
|
IFNA8
|
interferon alpha 8
|
| 3454
|
IFNAR1
|
interferon alpha and beta receptor subunit 1
|
| 3455
|
IFNAR2
|
interferon alpha and beta receptor subunit 2
|
| 338376
|
IFNE
|
interferon epsilon
|
| 116465
|
IFNGR1
|
interferon gamma receptor 1
|
| 3460
|
IFNGR2
|
interferon gamma receptor 2
|
| 56832
|
IFNK
|
interferon kappa
|
| 3467
|
IFNW1
|
interferon omega 1
|
| 25325
|
IL10
|
interleukin 10
|
| 3587
|
IL10RA
|
interleukin 10 receptor subunit alpha
|
| 3588
|
IL10RB
|
interleukin 10 receptor subunit beta
|
| 3592
|
IL12A
|
interleukin 12A
|
| 3597
|
IL13RA1
|
interleukin 13 receptor subunit alpha 1
|
| 25670
|
IL15
|
interleukin 15
|
| 3601
|
IL15RA
|
interleukin 15 receptor subunit alpha
|
| 29949
|
IL19
|
interleukin 19
|
| 50604
|
IL20
|
interleukin 20
|
| 53832
|
IL20RA
|
interleukin 20 receptor subunit alpha
|
| 53833
|
IL20RB
|
interleukin 20 receptor subunit beta
|
| 59067
|
IL21
|
interleukin 21
|
| 50615
|
IL21R
|
interleukin 21 receptor
|
| 50616
|
IL22
|
interleukin 22
|
| 58985
|
IL22RA1
|
interleukin 22 receptor subunit alpha 1
|
| 116379
|
IL22RA2
|
interleukin 22 receptor subunit alpha 2
|
| 11009
|
IL24
|
interleukin 24
|
| 55801
|
IL26
|
interleukin 26
|
| 282616
|
IL28A
|
interferon lambda 2
|
| 282617
|
IL28B
|
interferon lambda 3
|
| 163702
|
IL28RA
|
interferon lambda receptor 1
|
| 282618
|
IL29
|
interferon lambda 1
|
| 3563
|
IL3RA
|
interleukin 3 receptor subunit alpha
|
| 3566
|
IL4R
|
interleukin 4 receptor
|
| 3568
|
IL5RA
|
interleukin 5 receptor subunit alpha
|
| 3570
|
IL6R
|
interleukin 6 receptor
|
| 116558
|
IL7
|
interleukin 9
|
| 3575
|
IL7R
|
interleukin 7 receptor
|
| 3578
|
IL9
|
interleukin 9
|
| 3581
|
IL9R
|
interleukin 9 receptor
|
| 3953
|
LEPR
|
leptin receptor
|
| 3977
|
LIFR
|
LIF receptor alpha
|
| 9180
|
OSMR
|
oncostatin M receptor
|
| 85480
|
TSLP
|
thymic stromal lymphopoietin
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
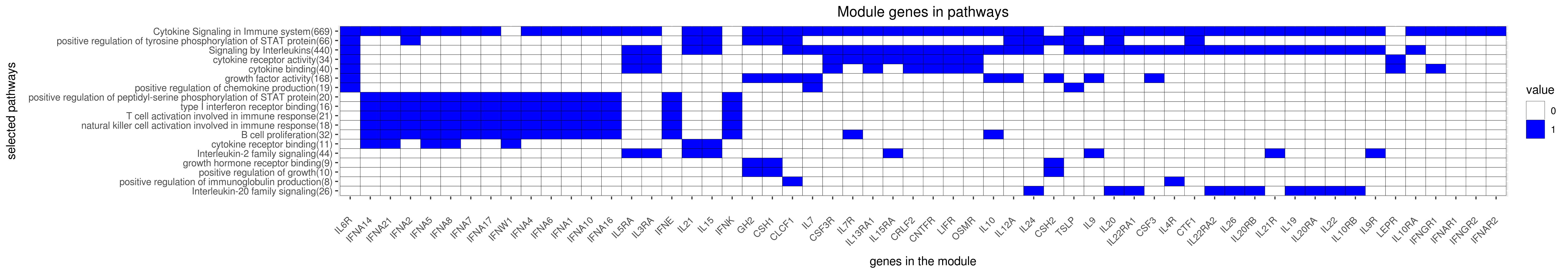
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0.00e+00
|
0.00e+00
|
TRAF6 MEDIATED IRF7 ACTIVATION
|
| 0.00e+00
|
0.00e+00
|
RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS
|
| 0.00e+00
|
0.00e+00
|
INTERFERON ALPHA BETA SIGNALING
|
| 0.00e+00
|
0.00e+00
|
REGULATION OF IFNA SIGNALING
|
| 0.00e+00
|
0.00e+00
|
FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION
|
| 0.00e+00
|
0.00e+00
|
RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS
|
| 0.00e+00
|
0.00e+00
|
INTERFERON ALPHA BETA SIGNALING
|
| 0.00e+00
|
0.00e+00
|
REGULATION OF IFNA SIGNALING
|
| 0.00e+00
|
0.00e+00
|
TRAF6 MEDIATED IRF7 ACTIVATION
|
| 0.00e+00
|
0.00e+00
|
CYTOKINE SIGNALING IN IMMUNE SYSTEM
|
| 0.00e+00
|
0.00e+00
|
INTERFERON SIGNALING
|
| 1.13e-16
|
4.47e-14
|
CYTOKINE SIGNALING IN IMMUNE SYSTEM
|
| 1.13e-12
|
4.44e-10
|
INTERFERON SIGNALING
|
| 2.76e-09
|
8.56e-07
|
FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION
|
| 4.73e-09
|
1.07e-06
|
INNATE IMMUNE SYSTEM
|
| 4.41e-06
|
8.90e-04
|
INNATE IMMUNE SYSTEM
|
| 4.35e-05
|
5.88e-03
|
SIGNALING BY ILS
|
| 3.07e-04
|
3.50e-02
|
IL 7 SIGNALING
|
| 6.50e-04
|
6.83e-02
|
REGULATION OF IFNG SIGNALING
|
| 1.82e-03
|
1.71e-01
|
PROLACTIN RECEPTOR SIGNALING
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:20:53 2018 - R2HTML