Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod24
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod24 |
| Module size |
31 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 325
|
APCS
|
amyloid P component, serum
|
| 335
|
APOA1
|
apolipoprotein A1
|
| 80350
|
APOA2
|
lipoprotein(a) like 2, pseudogene
|
| 337
|
APOA4
|
apolipoprotein A4
|
| 116519
|
APOA5
|
apolipoprotein A5
|
| 338
|
APOB
|
apolipoprotein B
|
| 341
|
APOC1
|
apolipoprotein C1
|
| 344
|
APOC2
|
apolipoprotein C2
|
| 347
|
APOD
|
apolipoprotein D
|
| 319
|
APOF
|
apolipoprotein F
|
| 1071
|
CETP
|
cholesteryl ester transfer protein
|
| 339390
|
CLEC4G
|
C-type lectin domain family 4 member G
|
| 1471
|
CST3
|
cystatin C
|
| 2934
|
GSN
|
gelsolin
|
| 55600
|
ITLN1
|
intelectin 1
|
| 9445
|
ITM2B
|
integral membrane protein 2B
|
| 3933
|
LCN1
|
lipocalin 1
|
| 4057
|
LTF
|
lactotransferrin
|
| 4069
|
LYZ
|
lysozyme
|
| 4240
|
MFGE8
|
milk fat globule-EGF factor 8 protein
|
| 4504
|
MT3
|
metallothionein 3
|
| 4547
|
MTTP
|
microsomal triglyceride transfer protein
|
| 4878
|
NPPA
|
natriuretic peptide A
|
| 54959
|
ODAM
|
odontogenic, ameloblast associated
|
| 5360
|
PLTP
|
phospholipid transfer protein
|
| 5950
|
RBP4
|
retinol binding protein 4
|
| 949
|
SCARB1
|
scavenger receptor class B member 1
|
| 6406
|
SEMG1
|
semenogelin 1
|
| 64220
|
STRA6
|
stimulated by retinoic acid 6
|
| 7045
|
TGFBI
|
transforming growth factor beta induced
|
| 7276
|
TTR
|
transthyretin
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
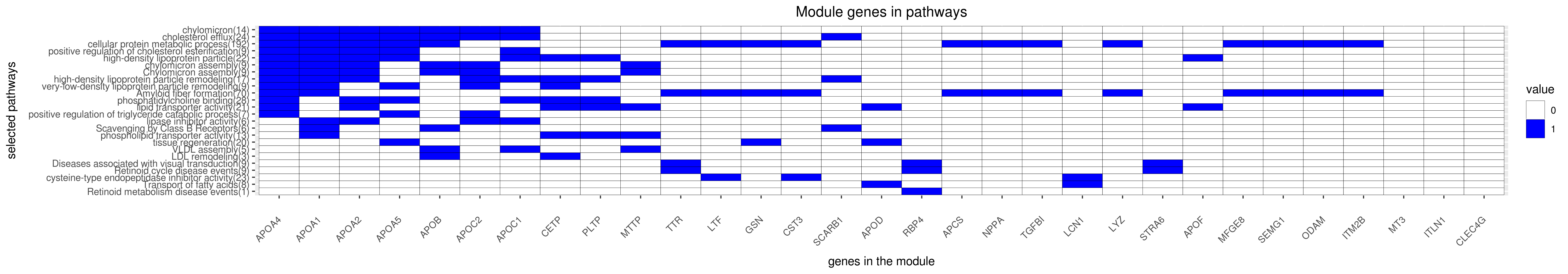
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0.00e+00
|
0.00e+00
|
LIPOPROTEIN METABOLISM
|
| 0.00e+00
|
0.00e+00
|
CHYLOMICRON MEDIATED LIPID TRANSPORT
|
| 0.00e+00
|
0.00e+00
|
LIPID DIGESTION MOBILIZATION AND TRANSPORT
|
| 0.00e+00
|
0.00e+00
|
AMYLOIDS
|
| 0.00e+00
|
0.00e+00
|
LIPOPROTEIN METABOLISM
|
| 0.00e+00
|
0.00e+00
|
LIPID DIGESTION MOBILIZATION AND TRANSPORT
|
| 0.00e+00
|
0.00e+00
|
AMYLOIDS
|
| 2.20e-12
|
6.49e-10
|
CHYLOMICRON MEDIATED LIPID TRANSPORT
|
| 2.73e-10
|
9.49e-08
|
HDL MEDIATED LIPID TRANSPORT
|
| 9.07e-10
|
2.22e-07
|
HDL MEDIATED LIPID TRANSPORT
|
| 1.42e-03
|
1.59e-01
|
PPARA ACTIVATES GENE EXPRESSION
|
| 2.81e-03
|
2.50e-01
|
PPARA ACTIVATES GENE EXPRESSION
|
| 8.40e-03
|
6.98e-01
|
FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM
|
| 1.87e-02
|
1.00e+00
|
FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM
|
| 2.28e-02
|
1.00e+00
|
PLATELET SENSITIZATION BY LDL
|
| 2.70e-02
|
1.00e+00
|
PLATELET SENSITIZATION BY LDL
|
| 2.96e-02
|
1.00e+00
|
CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS
|
| 3.55e-02
|
1.00e+00
|
CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS
|
| 3.89e-02
|
1.00e+00
|
ABCA TRANSPORTERS IN LIPID HOMEOSTASIS
|
| 3.90e-02
|
1.00e+00
|
ABCA TRANSPORTERS IN LIPID HOMEOSTASIS
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:20:50 2018 - R2HTML