Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod232
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod232 |
| Module size |
34 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 10438
|
C1D
|
C1D nuclear receptor corepressor
|
| 100912393
|
DCP2
|
mRNA-decapping enzyme 2-like
|
| 22894
|
DIS3
|
DIS3 homolog, exosome endoribonuclease and 3'-5' exoribonuclease
|
| 115752
|
DIS3L
|
DIS3 like exosome 3'-5' exoribonuclease
|
| 129563
|
DIS3L2
|
DIS3 like 3'-5' exoribonuclease 2
|
| 679140
|
EXOSC1
|
exosome component 1
|
| 5394
|
EXOSC10
|
exosome component 10
|
| 23404
|
EXOSC2
|
exosome component 2
|
| 51010
|
EXOSC3
|
exosome component 3
|
| 54512
|
EXOSC4
|
exosome component 4
|
| 56915
|
EXOSC5
|
exosome component 5
|
| 118460
|
EXOSC6
|
exosome component 6
|
| 23016
|
EXOSC7
|
exosome component 7
|
| 11340
|
EXOSC8
|
exosome component 8
|
| 5393
|
EXOSC9
|
exosome component 9
|
| 54707
|
GPN2
|
GPN-loop GTPase 2
|
| 9567
|
GTPBP1
|
GTP binding protein 1
|
| 8570
|
KHSRP
|
KH-type splicing regulatory protein
|
| 114785
|
MBD6
|
methyl-CpG binding domain protein 6
|
| 10200
|
MPHOSPH6
|
M-phase phosphoprotein 6
|
| 5073
|
PARN
|
poly(A)-specific ribonuclease
|
| 315045
|
POP1
|
POP1 homolog, ribonuclease P/MRP subunit
|
| 286430
|
POP4
|
NLR family pyrin domain containing 2B
|
| 51367
|
POP5
|
POP5 homolog, ribonuclease P/MRP subunit
|
| 10248
|
POP7
|
POP7 homolog, ribonuclease P/MRP subunit
|
| 361799
|
RAI1
|
decapping exoribonuclease
|
| 11102
|
RPP14
|
ribonuclease P/MRP subunit p14
|
| 79897
|
RPP21
|
ribonuclease P/MRP subunit p21
|
| 54913
|
RPP25
|
ribonuclease P and MRP subunit p25
|
| 10556
|
RPP30
|
ribonuclease P/MRP subunit p30
|
| 10799
|
RPP40
|
ribonuclease P/MRP subunit p40
|
| 288615
|
SBDS
|
SBDS, ribosome maturation factor
|
| 6832
|
SUPV3L1
|
Suv3 like RNA helicase
|
| 54464
|
XRN1
|
5'-3' exoribonuclease 1
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
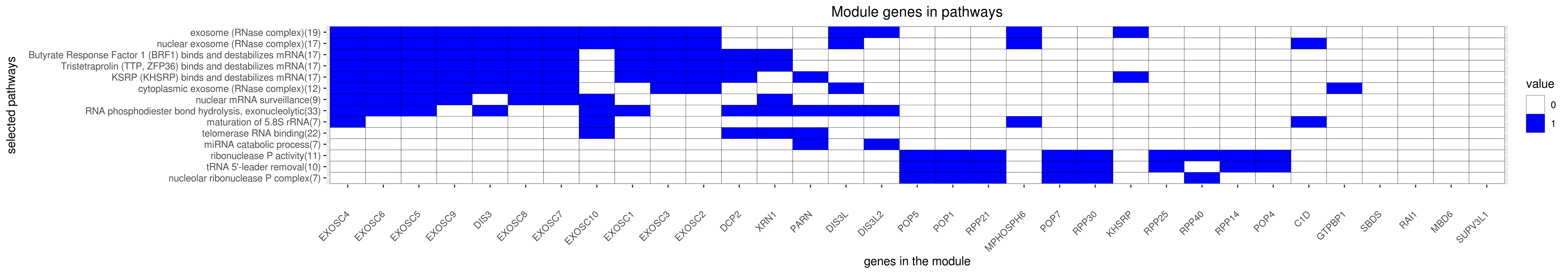
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0e+00
|
0e+00
|
DIABETES PATHWAYS
|
| 0e+00
|
0e+00
|
PERK REGULATED GENE EXPRESSION
|
| 0e+00
|
0e+00
|
UNFOLDED PROTEIN RESPONSE
|
| 0e+00
|
0e+00
|
MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE
|
| 0e+00
|
0e+00
|
REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS
|
| 0e+00
|
0e+00
|
DESTABILIZATION OF MRNA BY KSRP
|
| 0e+00
|
0e+00
|
ACTIVATION OF GENES BY ATF4
|
| 0e+00
|
0e+00
|
DEADENYLATION DEPENDENT MRNA DECAY
|
| 0e+00
|
0e+00
|
DESTABILIZATION OF MRNA BY BRF1
|
| 0e+00
|
0e+00
|
METABOLISM OF RNA
|
| 0e+00
|
0e+00
|
DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP
|
| 0e+00
|
0e+00
|
METABOLISM OF MRNA
|
| 0e+00
|
0e+00
|
ACTIVATION OF GENES BY ATF4
|
| 0e+00
|
0e+00
|
DIABETES PATHWAYS
|
| 0e+00
|
0e+00
|
UNFOLDED PROTEIN RESPONSE
|
| 0e+00
|
0e+00
|
PERK REGULATED GENE EXPRESSION
|
| 0e+00
|
0e+00
|
DEADENYLATION DEPENDENT MRNA DECAY
|
| 0e+00
|
0e+00
|
DESTABILIZATION OF MRNA BY BRF1
|
| 0e+00
|
0e+00
|
METABOLISM OF RNA
|
| 0e+00
|
0e+00
|
DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:31:12 2018 - R2HTML