Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod215
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod215 |
| Module size |
21 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 165
|
AEBP1
|
AE binding protein 1
|
| 56265
|
CPXM1
|
carboxypeptidase X, M14 family member 1
|
| 119587
|
CPXM2
|
carboxypeptidase X, M14 family member 2
|
| 8532
|
CPZ
|
carboxypeptidase Z
|
| 5045
|
FURIN
|
furin, paired basic amino acid cleaving enzyme
|
| 5122
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
| 27344
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor
|
| 5126
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
| 54760
|
PCSK4
|
proprotein convertase subtilisin/kexin type 4
|
| 5125
|
PCSK5
|
proprotein convertase subtilisin/kexin type 5
|
| 5046
|
PCSK6
|
proprotein convertase subtilisin/kexin type 6
|
| 9159
|
PCSK7
|
proprotein convertase subtilisin/kexin type 7
|
| 5545
|
PRB4
|
proline rich protein BstNI subfamily 4
|
| 284654
|
RSPO1
|
R-spondin 1
|
| 84870
|
RSPO3
|
R-spondin 3
|
| 343637
|
RSPO4
|
R-spondin 4
|
| 6447
|
SCG5
|
secretogranin V
|
| 23230
|
VPS13A
|
vacuolar protein sorting 13 homolog A
|
| 157680
|
VPS13B
|
vacuolar protein sorting 13 homolog B
|
| 54832
|
VPS13C
|
vacuolar protein sorting 13 homolog C
|
| 55187
|
VPS13D
|
vacuolar protein sorting 13 homolog D
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
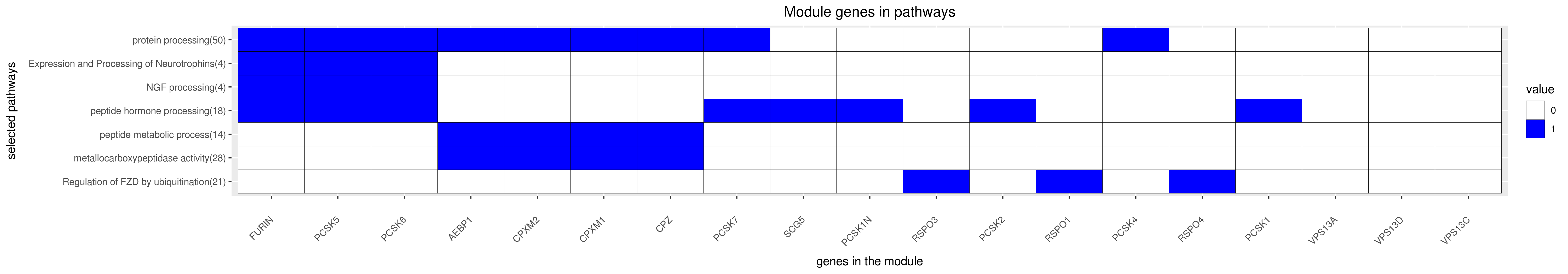
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 7.97e-05
|
1.27e-02
|
SIGNALLING BY NGF
|
| 9.23e-05
|
1.44e-02
|
SIGNALING BY NODAL
|
| 2.69e-04
|
3.72e-02
|
INSULIN SYNTHESIS AND PROCESSING
|
| 3.31e-04
|
3.75e-02
|
SIGNALING BY NODAL
|
| 3.66e-04
|
4.10e-02
|
INSULIN SYNTHESIS AND PROCESSING
|
| 2.07e-03
|
1.92e-01
|
SIGNALLING BY NGF
|
| 5.42e-03
|
4.39e-01
|
GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS
|
| 6.96e-03
|
5.45e-01
|
DIABETES PATHWAYS
|
| 7.22e-03
|
5.62e-01
|
PEPTIDE HORMONE BIOSYNTHESIS
|
| 7.76e-03
|
6.55e-01
|
DIABETES PATHWAYS
|
| 1.30e-02
|
9.80e-01
|
PEPTIDE HORMONE BIOSYNTHESIS
|
| 1.34e-02
|
1.00e+00
|
GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS
|
| 1.46e-02
|
9.94e-01
|
PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION
|
| 1.49e-02
|
1.00e+00
|
SYNTHESIS SECRETION AND INACTIVATION OF GIP
|
| 1.53e-02
|
1.00e+00
|
TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS
|
| 1.62e-02
|
1.00e+00
|
SYNTHESIS SECRETION AND INACTIVATION OF GIP
|
| 1.80e-02
|
1.00e+00
|
PRE NOTCH PROCESSING IN GOLGI
|
| 1.83e-02
|
1.00e+00
|
SYNTHESIS SECRETION AND INACTIVATION OF GLP1
|
| 1.94e-02
|
1.00e+00
|
INCRETIN SYNTHESIS SECRETION AND INACTIVATION
|
| 2.22e-02
|
1.00e+00
|
PRE NOTCH PROCESSING IN GOLGI
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:30:23 2018 - R2HTML