Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod211
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod211 |
| Module size |
30 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 392509
|
ARL13A
|
ADP ribosylation factor like GTPase 13A
|
| 402
|
ARL2
|
ADP ribosylation factor like GTPase 2
|
| 7461
|
CLIP2
|
CAP-Gly domain containing linker protein 2
|
| 28955
|
DEXI
|
Dexi homolog
|
| 5202
|
PFDN2
|
prefoldin subunit 2
|
| 5203
|
PFDN4
|
prefoldin subunit 4
|
| 10471
|
PFDN6
|
prefoldin subunit 6
|
| 100361193
|
TBCA
|
-
|
| 1155
|
TBCB
|
tubulin folding cofactor B
|
| 116653
|
TBCC
|
cyclase associated actin cytoskeleton regulatory protein 2
|
| 6904
|
TBCD
|
tubulin folding cofactor D
|
| 6905
|
TBCE
|
tubulin folding cofactor E
|
| 122664
|
TPPP2
|
tubulin polymerization promoting protein family member 2
|
| 10376
|
TUBA1B
|
tubulin alpha 1b
|
| 84790
|
TUBA1C
|
tubulin alpha 1c
|
| 7278
|
TUBA3C
|
tubulin alpha 3c
|
| 113457
|
TUBA3D
|
tubulin alpha 3d
|
| 112714
|
TUBA3E
|
tubulin alpha 3e
|
| 51807
|
TUBA8
|
tubulin alpha 8
|
| 79861
|
TUBAL3
|
tubulin alpha like 3
|
| 203068
|
TUBB
|
tubulin beta class I
|
| 81027
|
TUBB1
|
tubulin beta 1 class VI
|
| 7280
|
TUBB2A
|
tubulin beta 2A class IIa
|
| 347733
|
TUBB2B
|
tubulin beta 2B class IIb
|
| 10381
|
TUBB3
|
tubulin beta 3 class III
|
| 84617
|
TUBB6
|
tubulin beta 6 class V
|
| 347688
|
TUBB8
|
tubulin beta 8 class VIII
|
| 51174
|
TUBD1
|
tubulin delta 1
|
| 51175
|
TUBE1
|
tubulin epsilon 1
|
| 7411
|
VBP1
|
VHL binding protein 1
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
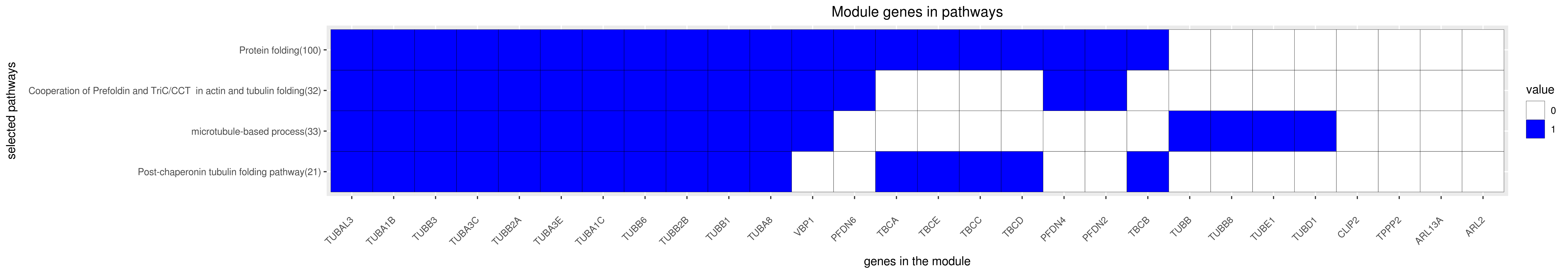
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0e+00
|
0e+00
|
POST CHAPERONIN TUBULIN FOLDING PATHWAY
|
| 0e+00
|
0e+00
|
PROTEIN FOLDING
|
| 0e+00
|
0e+00
|
PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC
|
| 0e+00
|
0e+00
|
FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC
|
| 0e+00
|
0e+00
|
POST CHAPERONIN TUBULIN FOLDING PATHWAY
|
| 0e+00
|
0e+00
|
PROTEIN FOLDING
|
| 0e+00
|
0e+00
|
PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC
|
| 0e+00
|
0e+00
|
FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:30:11 2018 - R2HTML