Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod193
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod193 |
| Module size |
31 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 1636
|
ACE
|
angiotensin I converting enzyme
|
| 59272
|
ACE2
|
angiotensin I converting enzyme 2
|
| 632
|
BGLAP
|
bone gamma-carboxyglutamate protein
|
| 671
|
BPI
|
bactericidal permeability increasing protein
|
| 389941
|
C1QL3
|
complement C1q like 3
|
| 1473
|
CST5
|
cystatin D
|
| 8530
|
CST7
|
cystatin F
|
| 1475
|
CSTA
|
cystatin A
|
| 1476
|
CSTB
|
cystatin B
|
| 1508
|
CTSB
|
cathepsin B
|
| 1075
|
CTSC
|
cathepsin C
|
| 1512
|
CTSH
|
cathepsin H
|
| 1513
|
CTSK
|
cathepsin K
|
| 1514
|
CTSL1
|
cathepsin L
|
| 1515
|
CTSL2
|
cathepsin V
|
| 1513
|
CTSO
|
cathepsin K
|
| 1520
|
CTSS
|
cathepsin S
|
| 1522
|
CTSZ
|
cathepsin Z
|
| 2896
|
GRN
|
granulin precursor
|
| 9452
|
ITM2A
|
integral membrane protein 2A
|
| 81618
|
ITM2C
|
integral membrane protein 2C
|
| 56660
|
KCNK12
|
potassium two pore domain channel subfamily K member 12
|
| 390940
|
PINLYP
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing
|
| 5972
|
REN
|
renin
|
| 347735
|
SERINC2
|
serine incorporator 2
|
| 871
|
SERPINH1
|
serpin family H member 1
|
| 142680
|
SLC34A3
|
solute carrier family 34 member 3
|
| 6590
|
SLPI
|
secretory leukocyte peptidase inhibitor
|
| 6692
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
| 6768
|
ST14
|
suppression of tumorigenicity 14
|
| 81622
|
UNC93B1
|
unc-93 homolog B1, TLR signaling regulator
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
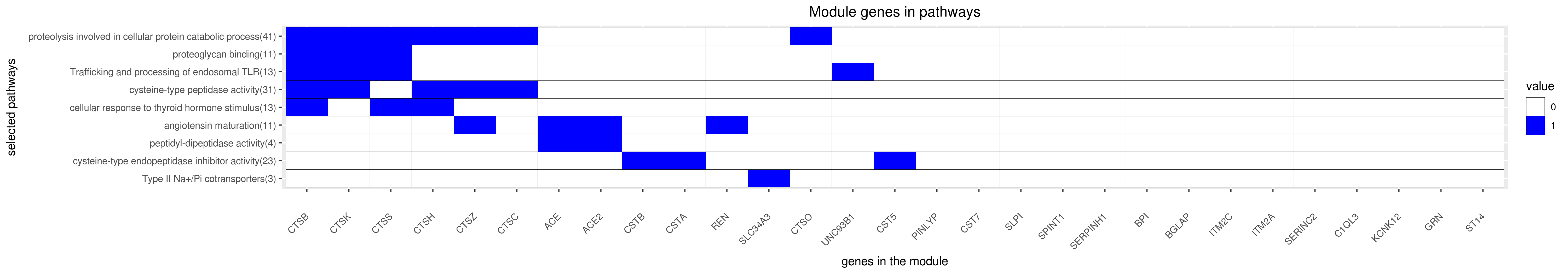
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0.00e+00
|
0.00e+00
|
MHC CLASS II ANTIGEN PRESENTATION
|
| 1.12e-11
|
3.27e-09
|
TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR
|
| 4.78e-11
|
1.78e-08
|
MHC CLASS II ANTIGEN PRESENTATION
|
| 2.58e-10
|
9.01e-08
|
TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR
|
| 1.48e-08
|
3.19e-06
|
TOLL RECEPTOR CASCADES
|
| 3.22e-08
|
8.86e-06
|
TOLL RECEPTOR CASCADES
|
| 9.81e-06
|
1.46e-03
|
INNATE IMMUNE SYSTEM
|
| 2.11e-05
|
3.79e-03
|
INNATE IMMUNE SYSTEM
|
| 6.86e-03
|
5.92e-01
|
ENDOSOMAL VACUOLAR PATHWAY
|
| 1.38e-02
|
9.51e-01
|
ENDOSOMAL VACUOLAR PATHWAY
|
| 1.99e-02
|
1.00e+00
|
LYSOSOME VESICLE BIOGENESIS
|
| 4.21e-02
|
1.00e+00
|
ANTIGEN PROCESSING CROSS PRESENTATION
|
| 4.29e-02
|
1.00e+00
|
LYSOSOME VESICLE BIOGENESIS
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:29:20 2018 - R2HTML