Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod188
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod188 |
| Module size |
41 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 150709
|
ANKAR
|
ankyrin and armadillo repeat containing
|
| 114898
|
C1QTNF2
|
C1q and TNF related 2
|
| 10518
|
CIB2
|
calcium and integrin binding family member 2
|
| 117286
|
CIB3
|
calcium and integrin binding family member 3
|
| 83716
|
CRISPLD2
|
cysteine rich secretory protein LCCL domain containing 2
|
| 1523
|
CUX1
|
cut like homeobox 1
|
| 23316
|
CUX2
|
cut like homeobox 2
|
| 79645
|
EFCAB1
|
EF-hand calcium binding domain 1
|
| 146330
|
FBXL16
|
F-box and leucine rich repeat protein 16
|
| 55612
|
FERMT1
|
fermitin family member 1
|
| 3208
|
HPCA
|
hippocalcin
|
| 3241
|
HPCAL1
|
hippocalcin like 1
|
| 51440
|
HPCAL4
|
hippocalcin like 4
|
| 11141
|
IL1RAPL1
|
interleukin 1 receptor accessory protein like 1
|
| 9657
|
IQCB1
|
IQ motif containing B1
|
| 10788
|
IQGAP2
|
IQ motif containing GTPase activating protein 2
|
| 128239
|
IQGAP3
|
IQ motif containing GTPase activating protein 3
|
| 30820
|
KCNIP1
|
potassium voltage-gated channel interacting protein 1
|
| 30819
|
KCNIP2
|
potassium voltage-gated channel interacting protein 2
|
| 30818
|
KCNIP3
|
potassium voltage-gated channel interacting protein 3
|
| 80333
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4
|
| 23026
|
MYO16
|
myosin XVI
|
| 80179
|
MYO19
|
myosin XIX
|
| 4640
|
MYO1A
|
myosin IA
|
| 4430
|
MYO1B
|
myosin IB
|
| 4642
|
MYO1D
|
myosin ID
|
| 4643
|
MYO1E
|
myosin IE
|
| 4542
|
MYO1F
|
myosin IF
|
| 64005
|
MYO1G
|
myosin IG
|
| 283446
|
MYO1H
|
myosin IH
|
| 4646
|
MYO6
|
myosin VI
|
| 23413
|
NCS1
|
neuronal calcium sensor 1
|
| 654790
|
PCP4L1
|
Purkinje cell protein 4 like 1
|
| 54922
|
RASIP1
|
Ras interacting protein 1
|
| 1827
|
RCAN1
|
regulator of calcineurin 1
|
| 10231
|
RCAN2
|
regulator of calcineurin 2
|
| 11123
|
RCAN3
|
RCAN family member 3
|
| 357
|
SHROOM2
|
shroom family member 2
|
| 55898
|
UNC45A
|
unc-45 myosin chaperone A
|
| 146862
|
UNC45B
|
unc-45 myosin chaperone B
|
| 7447
|
VSNL1
|
visinin like 1
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
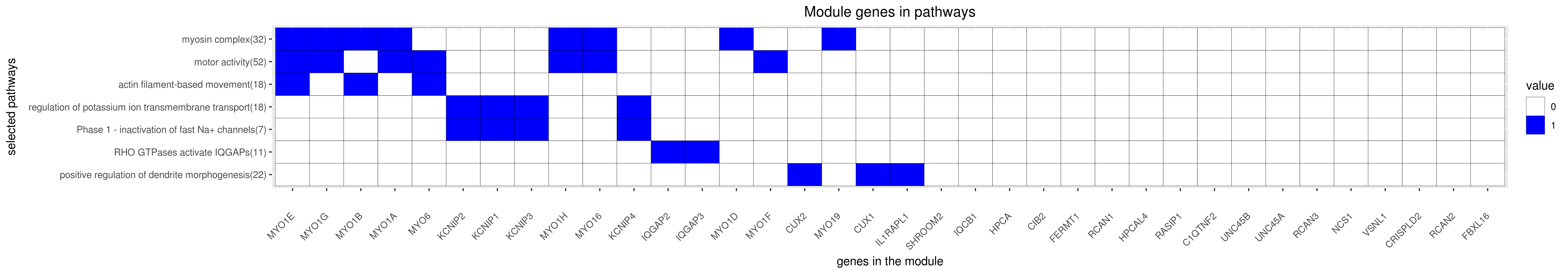
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 3.42e-03
|
2.97e-01
|
GAP JUNCTION DEGRADATION
|
| 3.44e-03
|
3.35e-01
|
GAP JUNCTION DEGRADATION
|
| 5.16e-03
|
4.69e-01
|
TRAFFICKING OF AMPA RECEPTORS
|
| 6.05e-03
|
5.35e-01
|
SIGNALING BY FGFR1 FUSION MUTANTS
|
| 6.06e-03
|
5.36e-01
|
SIGNALING BY FGFR1 MUTANTS
|
| 6.07e-03
|
5.37e-01
|
SIGNALING BY FGFR MUTANTS
|
| 8.25e-03
|
6.28e-01
|
GAP JUNCTION TRAFFICKING
|
| 8.29e-03
|
6.90e-01
|
SIGNALING BY FGFR IN DISEASE
|
| 1.50e-02
|
1.00e+00
|
SIGNALING BY FGFR1 MUTANTS
|
| 1.50e-02
|
1.00e+00
|
SIGNALING BY FGFR1 FUSION MUTANTS
|
| 1.51e-02
|
1.00e+00
|
SIGNALING BY FGFR MUTANTS
|
| 1.85e-02
|
1.00e+00
|
SIGNALING BY FGFR IN DISEASE
|
| 2.05e-02
|
1.00e+00
|
TRAFFICKING OF AMPA RECEPTORS
|
| 3.09e-02
|
1.00e+00
|
NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL
|
| 3.33e-02
|
1.00e+00
|
GAP JUNCTION TRAFFICKING
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:29:06 2018 - R2HTML