Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod186
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod186 |
| Module size |
21 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 84226
|
C2orf16
|
chromosome 2 open reading frame 16
|
| 1400
|
CRMP1
|
collapsin response mediator protein 1
|
| 51706
|
CYB5R1
|
cytochrome b5 reductase 1
|
| 51700
|
CYB5R2
|
cytochrome b5 reductase 2
|
| 1727
|
CYB5R3
|
cytochrome b5 reductase 3
|
| 51167
|
CYB5R4
|
cytochrome b5 reductase 4
|
| 606495
|
CYB5RL
|
cytochrome b5 reductase like
|
| 1723
|
DHODH
|
dihydroorotate dehydrogenase (quinone)
|
| 1806
|
DPYD
|
dihydropyrimidine dehydrogenase
|
| 1807
|
DPYS
|
dihydropyrimidinase
|
| 1808
|
DPYSL2
|
dihydropyrimidinase like 2
|
| 1809
|
DPYSL3
|
dihydropyrimidinase like 3
|
| 10570
|
DPYSL4
|
dihydropyrimidinase like 4
|
| 56896
|
DPYSL5
|
dihydropyrimidinase like 5
|
| 2232
|
FDXR
|
ferredoxin reductase
|
| 83943
|
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2
|
| 92106
|
OXNAD1
|
oxidoreductase NAD binding domain containing 1
|
| 9963
|
SLC23A1
|
solute carrier family 23 member 1
|
| 9962
|
SLC23A2
|
solute carrier family 23 member 2
|
| 151295
|
SLC23A3
|
solute carrier family 23 member 3
|
| 7372
|
UMPS
|
uridine monophosphate synthetase
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
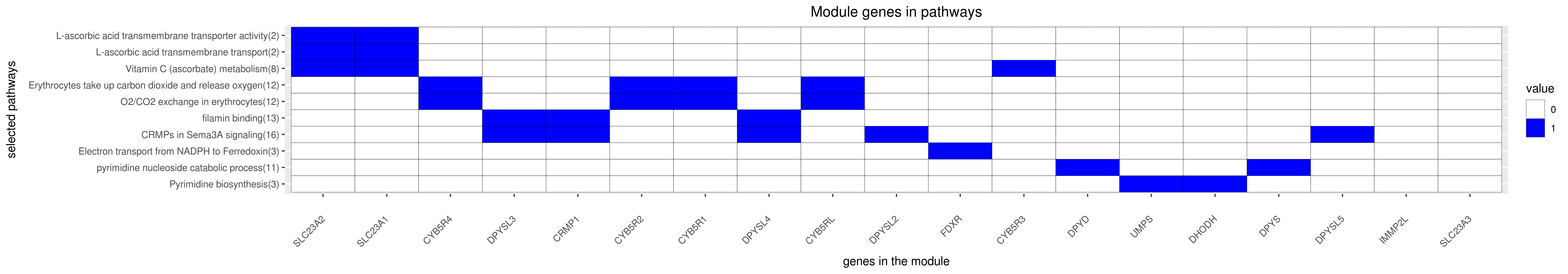
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 3.06e-11
|
1.15e-08
|
CRMPS IN SEMA3A SIGNALING
|
| 1.06e-10
|
2.89e-08
|
CRMPS IN SEMA3A SIGNALING
|
| 8.30e-09
|
2.46e-06
|
SEMAPHORIN INTERACTIONS
|
| 7.37e-08
|
1.46e-05
|
SEMAPHORIN INTERACTIONS
|
| 2.41e-07
|
5.94e-05
|
PYRIMIDINE METABOLISM
|
| 3.33e-07
|
6.15e-05
|
PYRIMIDINE METABOLISM
|
| 9.21e-06
|
1.77e-03
|
AXON GUIDANCE
|
| 2.87e-05
|
5.05e-03
|
METABOLISM OF NUCLEOTIDES
|
| 4.18e-05
|
5.68e-03
|
AXON GUIDANCE
|
| 5.41e-05
|
7.24e-03
|
METABOLISM OF NUCLEOTIDES
|
| 1.05e-04
|
1.62e-02
|
METABOLISM OF VITAMINS AND COFACTORS
|
| 1.34e-04
|
2.02e-02
|
DEVELOPMENTAL BIOLOGY
|
| 3.15e-04
|
4.29e-02
|
PYRIMIDINE CATABOLISM
|
| 6.71e-04
|
7.04e-02
|
METABOLISM OF VITAMINS AND COFACTORS
|
| 8.49e-04
|
8.68e-02
|
PYRIMIDINE CATABOLISM
|
| 1.85e-02
|
1.00e+00
|
RECYCLING PATHWAY OF L1
|
| 2.21e-02
|
1.00e+00
|
RECYCLING PATHWAY OF L1
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:28:59 2018 - R2HTML