Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod185
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod185 |
| Module size |
26 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 869
|
CBLN1
|
cerebellin 1 precursor
|
| 147381
|
CBLN2
|
cerebellin 2 precursor
|
| 643866
|
CBLN3
|
cerebellin 3 precursor
|
| 140689
|
CBLN4
|
cerebellin 4 precursor
|
| 1080
|
CFTR
|
cystic fibrosis transmembrane conductance regulator
|
| 1193
|
CLIC2
|
chloride intracellular channel 2
|
| 25932
|
CLIC4
|
chloride intracellular channel 4
|
| 53405
|
CLIC5
|
chloride intracellular channel 5
|
| 54102
|
CLIC6
|
chloride intracellular channel 6
|
| 286464
|
CXorf22
|
cilia and flagella associated protein 47
|
| 10243
|
GPHN
|
gephyrin
|
| 374918
|
IGFL1
|
IGF like family member 1
|
| 388555
|
IGFL3
|
IGF like family member 3
|
| 81689
|
ISCA1
|
iron-sulfur cluster assembly 1
|
| 122961
|
ISCA2
|
iron-sulfur cluster assembly 2
|
| 55034
|
MOCOS
|
molybdenum cofactor sulfurase
|
| 4810
|
NHS
|
NHS actin remodeling regulator
|
| 5023
|
P2RX1
|
purinergic receptor P2X 1
|
| 22953
|
P2RX2
|
purinergic receptor P2X 2
|
| 5024
|
P2RX3
|
purinergic receptor P2X 3
|
| 5334
|
PLCL1
|
phospholipase C like 1 (inactive)
|
| 54870
|
QRICH1
|
glutamine rich 1
|
| 92400
|
RBM18
|
RNA binding motif protein 18
|
| 221409
|
SPATS1
|
spermatogenesis associated serine rich 1
|
| 56674
|
TMEM9B
|
TMEM9 domain family member B
|
| 4308
|
TRPM1
|
transient receptor potential cation channel subfamily M member 1
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
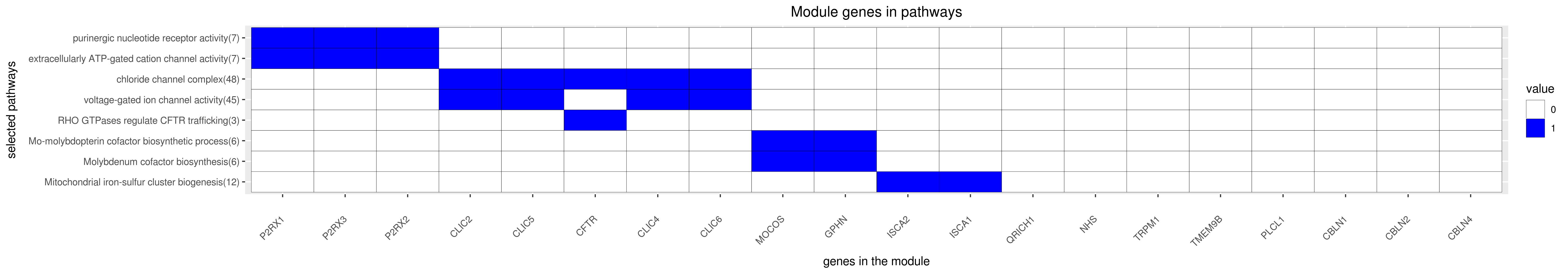
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 8.37e-04
|
8.58e-02
|
ELEVATION OF CYTOSOLIC CA2 LEVELS
|
| 8.38e-04
|
8.58e-02
|
PLATELET CALCIUM HOMEOSTASIS
|
| 2.78e-03
|
2.81e-01
|
METABOLISM OF VITAMINS AND COFACTORS
|
| 3.29e-03
|
3.25e-01
|
ELEVATION OF CYTOSOLIC CA2 LEVELS
|
| 3.30e-03
|
3.25e-01
|
PLATELET CALCIUM HOMEOSTASIS
|
| 7.60e-03
|
5.86e-01
|
PLATELET HOMEOSTASIS
|
| 8.07e-03
|
6.16e-01
|
METABOLISM OF VITAMINS AND COFACTORS
|
| 1.32e-02
|
9.91e-01
|
PLATELET HOMEOSTASIS
|
| 2.93e-02
|
1.00e+00
|
ABC FAMILY PROTEINS MEDIATED TRANSPORT
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:28:56 2018 - R2HTML