Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod176
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod176 |
| Module size |
26 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 64625
|
BID
|
BH3 interacting domain death agonist
|
| 329
|
BIRC2
|
baculoviral IAP repeat containing 2
|
| 330
|
BIRC3
|
baculoviral IAP repeat containing 3
|
| 843
|
CASP10
|
caspase 10
|
| 835
|
CASP2
|
caspase 2
|
| 836
|
CASP3
|
caspase 3
|
| 841
|
CASP8
|
caspase 8
|
| 8837
|
CFLAR
|
CASP8 and FADD like apoptosis regulator
|
| 8738
|
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain
|
| 9191
|
DEDD
|
death effector domain containing
|
| 8772
|
FADD
|
Fas associated via death domain
|
| 2194
|
FAS
|
fatty acid synthase
|
| 356
|
FASLG
|
Fas ligand
|
| 2108
|
MADD
|
electron transfer flavoprotein subunit alpha
|
| 9448
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
| 8996
|
NOL3
|
nucleolar protein 3
|
| 8439
|
NSMAF
|
neutral sphingomyelinase activation associated factor
|
| 389643
|
NUGGC
|
nuclear GTPase, germinal center associated
|
| 8797
|
TNFRSF10A
|
TNF receptor superfamily member 10a
|
| 8795
|
TNFRSF10B
|
TNF receptor superfamily member 10b
|
| 8794
|
TNFRSF10C
|
TNF receptor superfamily member 10c
|
| 8793
|
TNFRSF10D
|
TNF receptor superfamily member 10d
|
| 7132
|
TNFRSF1A
|
TNF receptor superfamily member 1A
|
| 8718
|
TNFRSF25
|
TNF receptor superfamily member 25
|
| 8743
|
TNFSF10
|
TNF superfamily member 10
|
| 8717
|
TRADD
|
TNFRSF1A associated via death domain
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
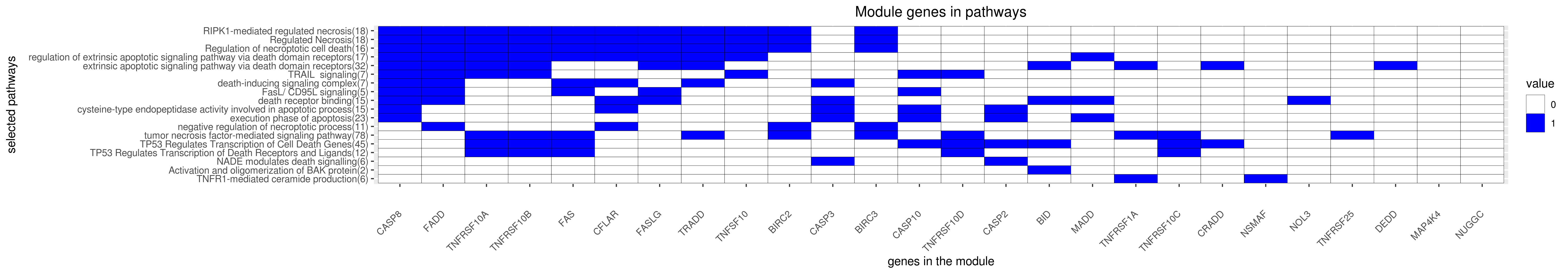
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0.00e+00
|
0.00e+00
|
EXTRINSIC PATHWAY FOR APOPTOSIS
|
| 0.00e+00
|
0.00e+00
|
EXTRINSIC PATHWAY FOR APOPTOSIS
|
| 0.00e+00
|
0.00e+00
|
APOPTOSIS
|
| 1.44e-10
|
5.17e-08
|
APOPTOSIS
|
| 6.89e-07
|
1.23e-04
|
NOD1 2 SIGNALING PATHWAY
|
| 3.50e-06
|
5.61e-04
|
NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10
|
| 3.64e-06
|
5.81e-04
|
NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS
|
| 2.04e-05
|
2.91e-03
|
INNATE IMMUNE SYSTEM
|
| 8.51e-05
|
1.10e-02
|
APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS
|
| 1.54e-04
|
1.87e-02
|
INTRINSIC PATHWAY FOR APOPTOSIS
|
| 1.95e-04
|
2.80e-02
|
NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10
|
| 1.98e-04
|
2.35e-02
|
APOPTOTIC EXECUTION PHASE
|
| 3.99e-04
|
4.43e-02
|
CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS
|
| 4.34e-04
|
5.68e-02
|
NOD1 2 SIGNALING PATHWAY
|
| 7.45e-04
|
7.71e-02
|
RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS
|
| 9.87e-04
|
1.17e-01
|
NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS
|
| 2.44e-03
|
2.52e-01
|
APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS
|
| 3.45e-03
|
3.36e-01
|
CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS
|
| 4.38e-03
|
4.10e-01
|
APOPTOTIC EXECUTION PHASE
|
| 4.88e-03
|
4.49e-01
|
INTRINSIC PATHWAY FOR APOPTOSIS
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:28:28 2018 - R2HTML