Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod167
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod167 |
| Module size |
16 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 1135
|
CHRNA2
|
cholinergic receptor nicotinic alpha 2 subunit
|
| 1136
|
CHRNA3
|
cholinergic receptor nicotinic alpha 3 subunit
|
| 1137
|
CHRNA4
|
cholinergic receptor nicotinic alpha 4 subunit
|
| 1138
|
CHRNA5
|
cholinergic receptor nicotinic alpha 5 subunit
|
| 8973
|
CHRNA6
|
cholinergic receptor nicotinic alpha 6 subunit
|
| 1139
|
CHRNA7
|
cholinergic receptor nicotinic alpha 7 subunit
|
| 55584
|
CHRNA9
|
cholinergic receptor nicotinic alpha 9 subunit
|
| 1140
|
CHRNB1
|
cholinergic receptor nicotinic beta 1 subunit
|
| 1141
|
CHRNB2
|
cholinergic receptor nicotinic beta 2 subunit
|
| 1142
|
CHRNB3
|
cholinergic receptor nicotinic beta 3 subunit
|
| 1143
|
CHRNB4
|
cholinergic receptor nicotinic beta 4 subunit
|
| 1144
|
CHRND
|
cholinergic receptor nicotinic delta subunit
|
| 1145
|
CHRNE
|
cholinergic receptor nicotinic epsilon subunit
|
| 1146
|
CHRNG
|
cholinergic receptor nicotinic gamma subunit
|
| 116372
|
LYPD1
|
LY6/PLAUR domain containing 1
|
| 134957
|
STXBP5
|
syntaxin binding protein 5
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
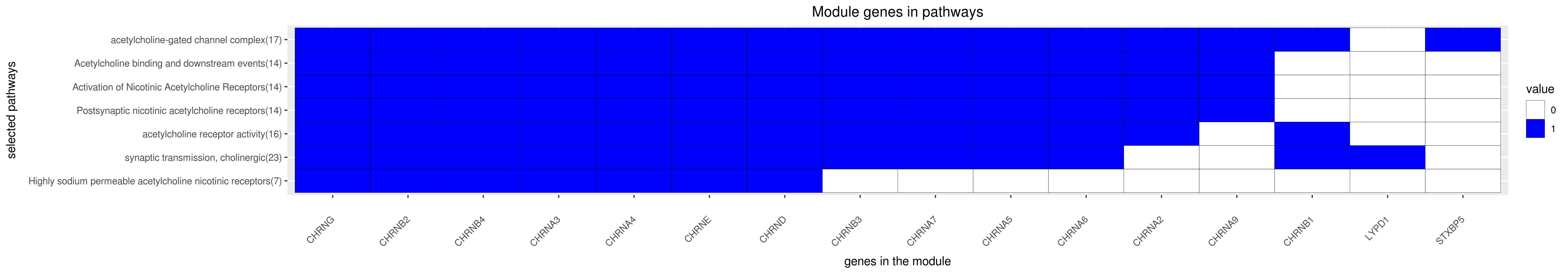
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0e+00
|
0e+00
|
PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS
|
| 0e+00
|
0e+00
|
NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL
|
| 0e+00
|
0e+00
|
ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS
|
| 0e+00
|
0e+00
|
HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS
|
| 0e+00
|
0e+00
|
NEURONAL SYSTEM
|
| 0e+00
|
0e+00
|
TRANSMISSION ACROSS CHEMICAL SYNAPSES
|
| 0e+00
|
0e+00
|
PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS
|
| 0e+00
|
0e+00
|
NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL
|
| 0e+00
|
0e+00
|
ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS
|
| 0e+00
|
0e+00
|
HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS
|
| 0e+00
|
0e+00
|
NEURONAL SYSTEM
|
| 0e+00
|
0e+00
|
TRANSMISSION ACROSS CHEMICAL SYNAPSES
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:28:01 2018 - R2HTML