Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod162
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod162 |
| Module size |
23 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 64431
|
ACTR6
|
ARP6 actin related protein 6 homolog
|
| 7862
|
BRPF1
|
bromodomain and PHD finger containing 1
|
| 10428
|
CFDP1
|
craniofacial development protein 1
|
| 298447
|
DMAP1
|
DNA methyltransferase 1-associated protein 1
|
| 57634
|
EP400
|
E1A binding protein p400
|
| 80314
|
EPC1
|
enhancer of polycomb homolog 1
|
| 26122
|
EPC2
|
enhancer of polycomb homolog 2
|
| 54556
|
ING3
|
inhibitor of growth family member 3
|
| 51147
|
ING4
|
inhibitor of growth family member 4
|
| 84289
|
ING5
|
inhibitor of growth family member 5
|
| 54617
|
INO80
|
INO80 complex subunit
|
| 64769
|
MEAF6
|
MYST/Esa1 associated factor 6
|
| 10933
|
MORF4L1
|
mortality factor 4 like 1
|
| 9643
|
MORF4L2
|
mortality factor 4 like 2
|
| 55257
|
MRGBP
|
MRG domain binding protein
|
| 10943
|
MSL3
|
MSL complex subunit 3
|
| 79960
|
PHF17
|
jade family PHD finger 1
|
| 8607
|
RUVBL1
|
RuvB like AAA ATPase 1
|
| 10856
|
RUVBL2
|
RuvB like AAA ATPase 2
|
| 10847
|
SRCAP
|
Snf2 related CREBBP activator protein
|
| 6944
|
VPS72
|
vacuolar protein sorting 72 homolog
|
| 8089
|
YEATS4
|
YEATS domain containing 4
|
| 10467
|
ZNHIT1
|
zinc finger HIT-type containing 1
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
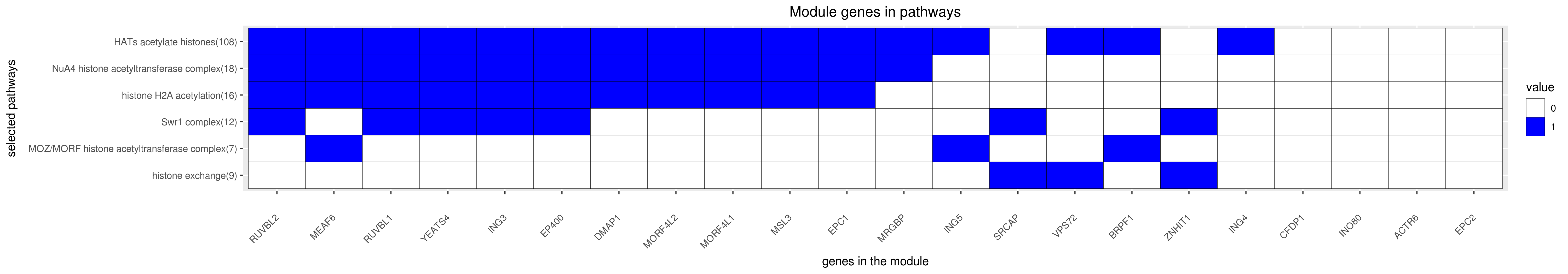
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 8.09e-06
|
1.23e-03
|
EXTENSION OF TELOMERES
|
| 8.46e-06
|
1.28e-03
|
TELOMERE MAINTENANCE
|
| 3.40e-05
|
5.90e-03
|
EXTENSION OF TELOMERES
|
| 6.25e-05
|
1.02e-02
|
TELOMERE MAINTENANCE
|
| 1.09e-04
|
1.38e-02
|
CHROMOSOME MAINTENANCE
|
| 4.70e-04
|
6.10e-02
|
CHROMOSOME MAINTENANCE
|
| 4.75e-03
|
4.38e-01
|
CELL CYCLE
|
| 4.88e-03
|
4.02e-01
|
DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE
|
| 1.03e-02
|
8.15e-01
|
DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:27:45 2018 - R2HTML