Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod155
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod155 |
| Module size |
22 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 1650
|
DDOST
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase non-catalytic subunit
|
| 6184
|
RPN1
|
ribophorin I
|
| 6185
|
RPN2
|
ribophorin II
|
| 23478
|
SEC11A
|
SEC11 homolog A, signal peptidase complex subunit
|
| 90701
|
SEC11C
|
SEC11 homolog C, signal peptidase complex subunit
|
| 10952
|
SEC61B
|
Sec61 translocon beta subunit
|
| 23480
|
SEC61G
|
Sec61 translocon gamma subunit
|
| 28972
|
SPCS1
|
signal peptidase complex subunit 1
|
| 9789
|
SPCS2
|
signal peptidase complex subunit 2
|
| 60559
|
SPCS3
|
signal peptidase complex subunit 3
|
| 296076
|
SRP14
|
signal recognition particle 14
|
| 291685
|
SRP19
|
signal recognition particle 19
|
| 116650
|
SRP54
|
signal recognition particle 54A
|
| 6730
|
SRP68
|
signal recognition particle 68
|
| 100909747
|
SRP72
|
signal recognition particle 72 kDa protein-like
|
| 6726
|
SRP9
|
signal recognition particle 9
|
| 6734
|
SRPR
|
SRP receptor subunit alpha
|
| 308821
|
SRPRB
|
RAB30, member RAS oncogene family
|
| 6745
|
SSR1
|
signal sequence receptor subunit 1
|
| 6746
|
SSR2
|
signal sequence receptor subunit 2
|
| 6747
|
SSR3
|
signal sequence receptor subunit 3
|
| 312903
|
TRAM1
|
translocation associated membrane protein 1
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
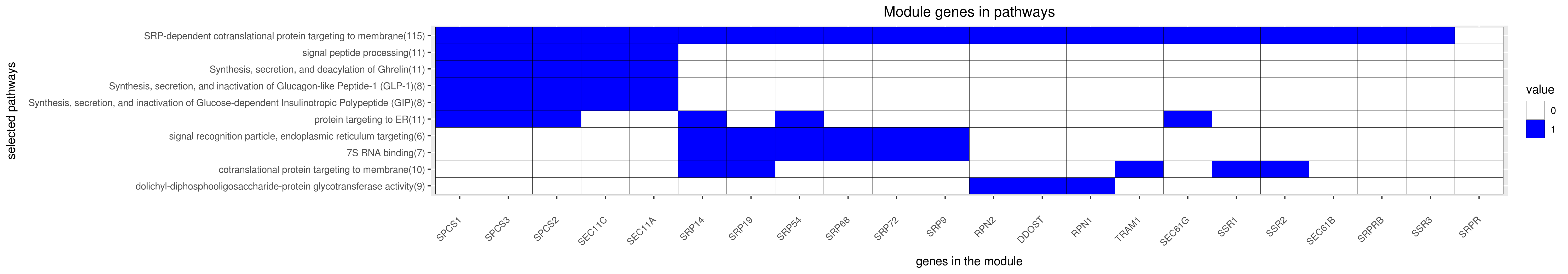
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0.00e+00
|
0.00e+00
|
SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE
|
| 0.00e+00
|
0.00e+00
|
TRANSLATION
|
| 0.00e+00
|
0.00e+00
|
SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE
|
| 0.00e+00
|
0.00e+00
|
TRANSLATION
|
| 4.28e-09
|
1.30e-06
|
SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN
|
| 4.30e-09
|
1.31e-06
|
SYNTHESIS SECRETION AND INACTIVATION OF GIP
|
| 4.35e-09
|
1.32e-06
|
DIABETES PATHWAYS
|
| 1.19e-08
|
3.45e-06
|
SYNTHESIS SECRETION AND INACTIVATION OF GLP1
|
| 1.40e-08
|
4.02e-06
|
INCRETIN SYNTHESIS SECRETION AND INACTIVATION
|
| 3.39e-08
|
7.03e-06
|
SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN
|
| 4.39e-08
|
8.94e-06
|
SYNTHESIS SECRETION AND INACTIVATION OF GIP
|
| 6.10e-08
|
1.22e-05
|
SYNTHESIS SECRETION AND INACTIVATION OF GLP1
|
| 1.33e-07
|
2.55e-05
|
DIABETES PATHWAYS
|
| 1.61e-07
|
3.07e-05
|
INCRETIN SYNTHESIS SECRETION AND INACTIVATION
|
| 2.45e-06
|
5.15e-04
|
REGULATION OF INSULIN SECRETION
|
| 8.29e-06
|
1.61e-03
|
INTEGRATION OF ENERGY METABOLISM
|
| 4.63e-05
|
6.22e-03
|
REGULATION OF INSULIN SECRETION
|
| 1.43e-04
|
1.76e-02
|
INTEGRATION OF ENERGY METABOLISM
|
| 6.20e-04
|
6.56e-02
|
ACTIVATION OF CHAPERONE GENES BY XBP1S
|
| 1.01e-03
|
1.19e-01
|
ACTIVATION OF CHAPERONE GENES BY XBP1S
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:27:24 2018 - R2HTML