Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod146
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod146 |
| Module size |
27 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 254427
|
C10orf47
|
proline and serine rich 2
|
| 51719
|
CAB39
|
calcium binding protein 39
|
| 81617
|
CAB39L
|
calcium binding protein 39 like
|
| 10129
|
FRY
|
FRY microtubule binding protein
|
| 83855
|
KLF16
|
Kruppel like factor 16
|
| 9113
|
LATS1
|
large tumor suppressor kinase 1
|
| 26524
|
LATS2
|
large tumor suppressor kinase 2
|
| 79705
|
LRRK1
|
leucine rich repeat kinase 1
|
| 1326
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
| 11184
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1
|
| 5871
|
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2
|
| 8491
|
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3
|
| 11183
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5
|
| 55233
|
MOB1A
|
MOB kinase activator 1A
|
| 92597
|
MOB1B
|
MOB kinase activator 1B
|
| 81532
|
MOB2
|
MOB kinase activator 2
|
| 126308
|
MOB3A
|
MOB kinase activator 3A
|
| 148932
|
MOB3C
|
MOB kinase activator 3C
|
| 56980
|
PRDM10
|
PR/SET domain 10
|
| 114821
|
SCAND3
|
zinc finger BED-type containing 9
|
| 8428
|
STK24
|
serine/threonine kinase 24
|
| 10494
|
STK25
|
serine/threonine kinase 25
|
| 8428
|
STK3
|
serine/threonine kinase 24
|
| 11329
|
STK38
|
serine/threonine kinase 38
|
| 23012
|
STK38L
|
serine/threonine kinase 38 like
|
| 6789
|
STK4
|
serine/threonine kinase 4
|
| 678
|
ZFP36L2
|
ZFP36 ring finger protein like 2
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
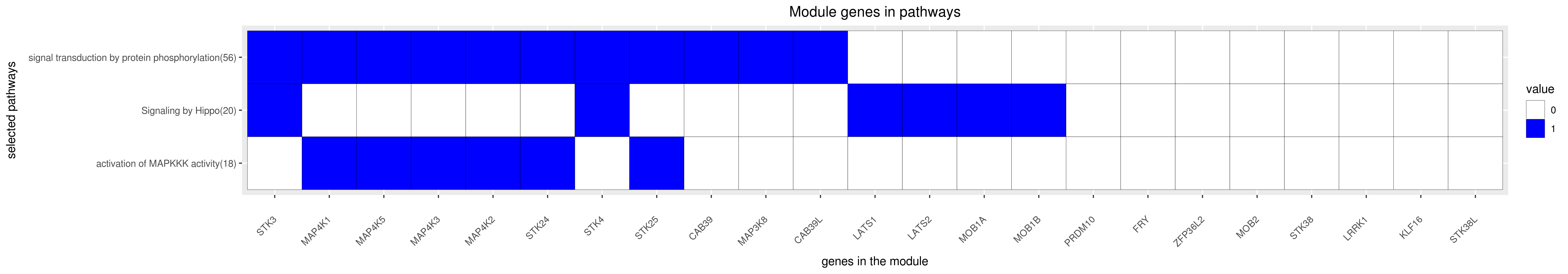
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0.00e+00
|
0.00e+00
|
SIGNALING BY HIPPO
|
| 7.12e-10
|
1.77e-07
|
SIGNALING BY HIPPO
|
| 2.83e-04
|
3.89e-02
|
REGULATION OF AMPK ACTIVITY VIA LKB1
|
| 4.11e-04
|
5.43e-02
|
ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK
|
| 5.54e-04
|
7.05e-02
|
ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE
|
| 6.86e-04
|
8.46e-02
|
PKB MEDIATED EVENTS
|
| 1.04e-03
|
1.04e-01
|
REGULATION OF AMPK ACTIVITY VIA LKB1
|
| 1.05e-03
|
1.05e-01
|
ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE
|
| 1.29e-03
|
1.26e-01
|
ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK
|
| 1.70e-03
|
1.86e-01
|
PI3K CASCADE
|
| 1.87e-03
|
1.75e-01
|
PKB MEDIATED EVENTS
|
| 2.01e-03
|
2.14e-01
|
INSULIN RECEPTOR SIGNALLING CASCADE
|
| 3.22e-03
|
2.83e-01
|
PI3K CASCADE
|
| 3.44e-03
|
3.35e-01
|
CD28 DEPENDENT PI3K AKT SIGNALING
|
| 3.80e-03
|
3.25e-01
|
INSULIN RECEPTOR SIGNALLING CASCADE
|
| 6.88e-03
|
5.92e-01
|
CD28 CO STIMULATION
|
| 7.34e-03
|
6.25e-01
|
SIGNALING BY INSULIN RECEPTOR
|
| 7.74e-03
|
5.95e-01
|
SIGNALING BY INSULIN RECEPTOR
|
| 9.04e-03
|
6.76e-01
|
CD28 DEPENDENT PI3K AKT SIGNALING
|
| 1.09e-02
|
7.84e-01
|
CD28 CO STIMULATION
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:26:56 2018 - R2HTML