Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod143
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod143 |
| Module size |
36 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 79738
|
BBS10
|
Bardet-Biedl syndrome 10
|
| 10576
|
CCT2
|
chaperonin containing TCP1 subunit 2
|
| 7203
|
CCT3
|
chaperonin containing TCP1 subunit 3
|
| 10575
|
CCT4
|
chaperonin containing TCP1 subunit 4
|
| 22948
|
CCT5
|
chaperonin containing TCP1 subunit 5
|
| 908
|
CCT6A
|
chaperonin containing TCP1 subunit 6A
|
| 10693
|
CCT6B
|
chaperonin containing TCP1 subunit 6B
|
| 10574
|
CCT7
|
chaperonin containing TCP1 subunit 7
|
| 10694
|
CCT8
|
chaperonin containing TCP1 subunit 8
|
| 150160
|
CCT8L2
|
chaperonin containing TCP1 subunit 8 like 2
|
| 55917
|
CTTNBP2NL
|
CTTNBP2 N-terminal like
|
| 653333
|
FAM86B2
|
family with sequence similarity 86 member B2
|
| 26127
|
FGFR1OP2
|
FGFR1 oncogene partner 2
|
| 3336
|
HSPE1
|
heat shock protein family E (Hsp10) member 1
|
| 92342
|
METTL18
|
methyltransferase like 18
|
| 25895
|
METTL21B
|
EEF1A lysine methyltransferase 3
|
| 8195
|
MKKS
|
McKusick-Kaufman syndrome
|
| 25843
|
MOB4
|
MOB family member 4, phocein
|
| 4973
|
OLR1
|
oxidized low density lipoprotein receptor 1
|
| 135138
|
PACRG
|
parkin coregulated
|
| 11235
|
PDCD10
|
programmed cell death 10
|
| 132954
|
PDCL2
|
phosducin like 2
|
| 79031
|
PDCL3
|
phosducin like 3
|
| 5201
|
PFDN1
|
prefoldin subunit 1
|
| 80143
|
SIKE1
|
suppressor of IKBKE 1
|
| 85369
|
STRIP1
|
striatin interacting protein 1
|
| 57464
|
STRIP2
|
striatin interacting protein 2
|
| 6801
|
STRN
|
striatin
|
| 29966
|
STRN3
|
striatin 3
|
| 29888
|
STRN4
|
striatin 4
|
| 6950
|
TCP1
|
t-complex 1
|
| 80342
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
| 118813
|
ZFYVE27
|
zinc finger FYVE-type containing 27
|
| 11055
|
ZPBP
|
zona pellucida binding protein
|
| 124626
|
ZPBP2
|
zona pellucida binding protein 2
|
| 54764
|
ZRANB1
|
zinc finger RANBP2-type containing 1
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
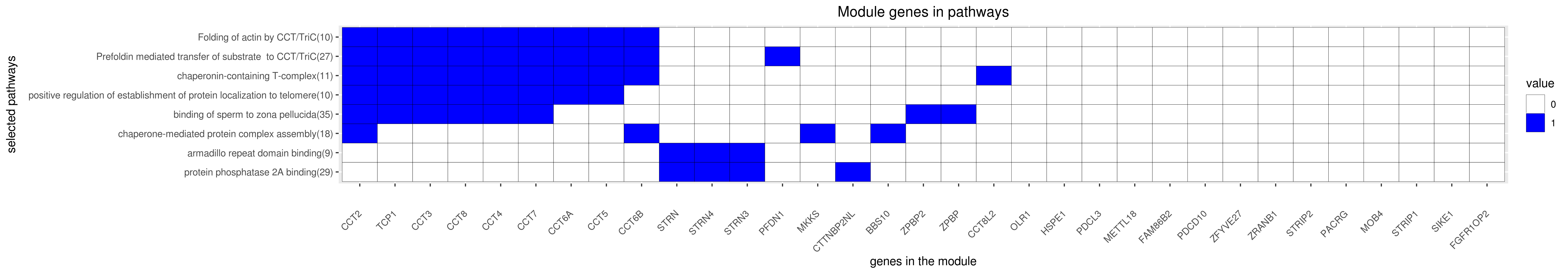
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0.00e+00
|
0.00e+00
|
PROTEIN FOLDING
|
| 0.00e+00
|
0.00e+00
|
PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC
|
| 0.00e+00
|
0.00e+00
|
ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS
|
| 0.00e+00
|
0.00e+00
|
FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC
|
| 0.00e+00
|
0.00e+00
|
PROTEIN FOLDING
|
| 0.00e+00
|
0.00e+00
|
PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC
|
| 0.00e+00
|
0.00e+00
|
ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS
|
| 0.00e+00
|
0.00e+00
|
FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC
|
| 3.30e-03
|
3.25e-01
|
TRAF3 DEPENDENT IRF ACTIVATION PATHWAY
|
| 3.30e-03
|
3.25e-01
|
TRAF6 MEDIATED IRF7 ACTIVATION
|
| 1.60e-02
|
1.00e+00
|
TRAF3 DEPENDENT IRF ACTIVATION PATHWAY
|
| 1.61e-02
|
1.00e+00
|
TRAF6 MEDIATED IRF7 ACTIVATION
|
| 1.81e-02
|
1.00e+00
|
RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS
|
| 4.78e-02
|
1.00e+00
|
RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:26:48 2018 - R2HTML