Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod142
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod142 |
| Module size |
32 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 375790
|
AGRN
|
agrin
|
| 126792
|
B3GALT6
|
beta-1,3-galactosyltransferase 6
|
| 11285
|
B4GALT7
|
beta-1,4-galactosyltransferase 7
|
| 2131
|
EXT1
|
exostosin glycosyltransferase 1
|
| 2132
|
EXT2
|
exostosin glycosyltransferase 2
|
| 26035
|
GLCE
|
glucuronic acid epimerase
|
| 221914
|
GPC2
|
glypican 2
|
| 2719
|
GPC3
|
glypican 3
|
| 2239
|
GPC4
|
glypican 4
|
| 2262
|
GPC5
|
glypican 5
|
| 10082
|
GPC6
|
glypican 6
|
| 283464
|
GXYLT1
|
glucoside xylosyltransferase 1
|
| 10855
|
HPSE
|
heparanase
|
| 60495
|
HPSE2
|
heparanase 2 (inactive)
|
| 9957
|
HS3ST1
|
heparan sulfate-glucosamine 3-sulfotransferase 1
|
| 9956
|
HS3ST2
|
heparan sulfate-glucosamine 3-sulfotransferase 2
|
| 9955
|
HS3ST3A1
|
heparan sulfate-glucosamine 3-sulfotransferase 3A1
|
| 9953
|
HS3ST3B1
|
heparan sulfate-glucosamine 3-sulfotransferase 3B1
|
| 9951
|
HS3ST4
|
heparan sulfate-glucosamine 3-sulfotransferase 4
|
| 64711
|
HS3ST5
|
heparan sulfate-glucosamine 3-sulfotransferase 6
|
| 64711
|
HS3ST6
|
heparan sulfate-glucosamine 3-sulfotransferase 6
|
| 9394
|
HS6ST1
|
heparan sulfate 6-O-sulfotransferase 1
|
| 90161
|
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2
|
| 3339
|
HSPG2
|
heparan sulfate proteoglycan 2
|
| 3425
|
IDUA
|
iduronidase, alpha-L-
|
| 3340
|
NDST1
|
N-deacetylase and N-sulfotransferase 1
|
| 8509
|
NDST2
|
N-deacetylase and N-sulfotransferase 2
|
| 6382
|
SDC1
|
syndecan 1
|
| 6383
|
SDC2
|
syndecan 2
|
| 9672
|
SDC3
|
syndecan 3
|
| 6385
|
SDC4
|
syndecan 4
|
| 11046
|
SLC35D2
|
solute carrier family 35 member D2
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
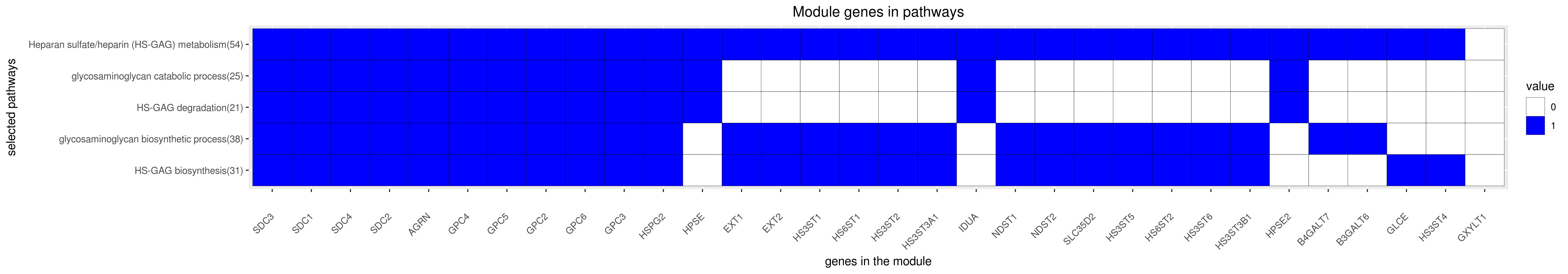
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0.00e+00
|
0.00e+00
|
HS GAG BIOSYNTHESIS
|
| 0.00e+00
|
0.00e+00
|
HEPARAN SULFATE HEPARIN HS GAG METABOLISM
|
| 0.00e+00
|
0.00e+00
|
GLYCOSAMINOGLYCAN METABOLISM
|
| 0.00e+00
|
0.00e+00
|
METABOLISM OF CARBOHYDRATES
|
| 0.00e+00
|
0.00e+00
|
A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS
|
| 0.00e+00
|
0.00e+00
|
HS GAG DEGRADATION
|
| 0.00e+00
|
0.00e+00
|
CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM
|
| 0.00e+00
|
0.00e+00
|
HS GAG BIOSYNTHESIS
|
| 0.00e+00
|
0.00e+00
|
HEPARAN SULFATE HEPARIN HS GAG METABOLISM
|
| 0.00e+00
|
0.00e+00
|
GLYCOSAMINOGLYCAN METABOLISM
|
| 0.00e+00
|
0.00e+00
|
METABOLISM OF CARBOHYDRATES
|
| 0.00e+00
|
0.00e+00
|
HS GAG DEGRADATION
|
| 0.00e+00
|
0.00e+00
|
A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS
|
| 0.00e+00
|
0.00e+00
|
CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM
|
| 7.77e-03
|
6.56e-01
|
CHYLOMICRON MEDIATED LIPID TRANSPORT
|
| 1.21e-02
|
8.59e-01
|
CHYLOMICRON MEDIATED LIPID TRANSPORT
|
| 2.06e-02
|
1.00e+00
|
LIPOPROTEIN METABOLISM
|
| 2.29e-02
|
1.00e+00
|
LIPOPROTEIN METABOLISM
|
| 3.55e-02
|
1.00e+00
|
LIPID DIGESTION MOBILIZATION AND TRANSPORT
|
| 3.90e-02
|
1.00e+00
|
LIPID DIGESTION MOBILIZATION AND TRANSPORT
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:26:45 2018 - R2HTML