Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod133
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod133 |
| Module size |
36 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 1602
|
DACH1
|
dachshund family transcription factor 1
|
| 117154
|
DACH2
|
dachshund family transcription factor 2
|
| 1745
|
DLX1
|
distal-less homeobox 1
|
| 1748
|
DLX4
|
distal-less homeobox 4
|
| 1750
|
DLX6
|
distal-less homeobox 6
|
| 22947
|
DUX4
|
double homeobox 4 like 1
|
| 728410
|
DUX4L2
|
double homeobox 4 like 2
|
| 653548
|
DUX4L3
|
double homeobox 4 like 3
|
| 441056
|
DUX4L4
|
double homeobox 4 like 4
|
| 653545
|
DUX4L5
|
double homeobox 4 like 5
|
| 653544
|
DUX4L6
|
double homeobox 4 like 6
|
| 653543
|
DUX4L7
|
double homeobox 4 like 7
|
| 503835
|
DUXA
|
double homeobox A
|
| 57593
|
EBF4
|
early B cell factor 4
|
| 2138
|
EYA1
|
EYA transcriptional coactivator and phosphatase 1
|
| 2139
|
EYA2
|
EYA transcriptional coactivator and phosphatase 2
|
| 2140
|
EYA3
|
EYA transcriptional coactivator and phosphatase 3
|
| 2070
|
EYA4
|
EYA transcriptional coactivator and phosphatase 4
|
| 3166
|
HMX1
|
H6 family homeobox 1
|
| 83881
|
MIXL1
|
Mix paired-like homeobox
|
| 5077
|
PAX3
|
paired box 3
|
| 5078
|
PAX4
|
paired box 4
|
| 5080
|
PAX6
|
paired box 6
|
| 5081
|
PAX7
|
paired box 7
|
| 6495
|
SIX1
|
SIX homeobox 1
|
| 10736
|
SIX2
|
SIX homeobox 2
|
| 6496
|
SIX3
|
SIX homeobox 3
|
| 51804
|
SIX4
|
SIX homeobox 4
|
| 147912
|
SIX5
|
SIX homeobox 5
|
| 4990
|
SIX6
|
SIX homeobox 6
|
| 9095
|
TBX19
|
T-box 19
|
| 6909
|
TBX2
|
T-box 2
|
| 9496
|
TBX4
|
T-box 4
|
| 11023
|
VAX1
|
ventral anterior homeobox 1
|
| 25806
|
VAX2
|
ventral anterior homeobox 2
|
| 338917
|
VSX2
|
visual system homeobox 2
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
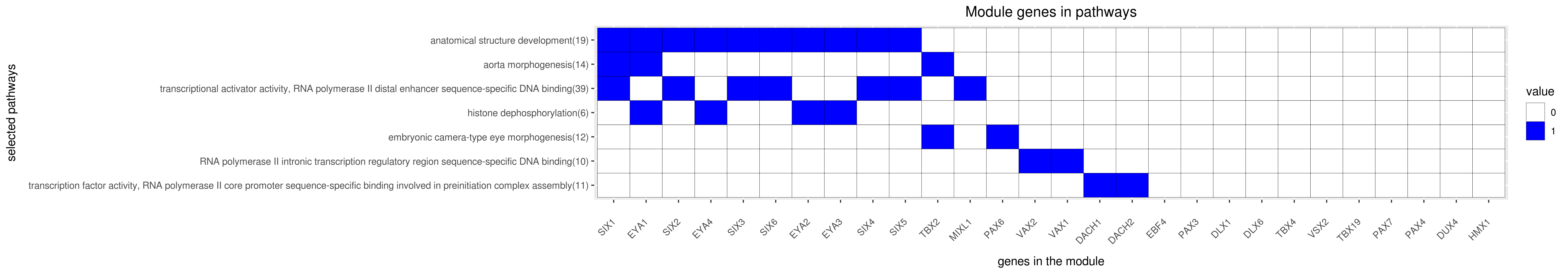
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 3.44e-03
|
3.35e-01
|
REGULATION OF GENE EXPRESSION IN BETA CELLS
|
| 5.05e-03
|
4.61e-01
|
REGULATION OF BETA CELL DEVELOPMENT
|
| 1.37e-02
|
1.00e+00
|
SYNTHESIS SECRETION AND INACTIVATION OF GIP
|
| 1.37e-02
|
1.00e+00
|
SYNTHESIS SECRETION AND INACTIVATION OF GLP1
|
| 1.37e-02
|
1.00e+00
|
INCRETIN SYNTHESIS SECRETION AND INACTIVATION
|
| 2.98e-02
|
1.00e+00
|
REGULATION OF INSULIN SECRETION
|
| 3.82e-02
|
1.00e+00
|
INTEGRATION OF ENERGY METABOLISM
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:26:17 2018 - R2HTML