Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod129
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod129 |
| Module size |
59 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 8092
|
ALX1
|
ALX homeobox 1
|
| 257
|
ALX3
|
ALX homeobox 3
|
| 60529
|
ALX4
|
ALX homeobox 4
|
| 10054
|
ARX
|
ubiquitin like modifier activating enzyme 2
|
| 343472
|
BARHL2
|
BarH like homeobox 2
|
| 120237
|
DBX1
|
developing brain homeobox 1
|
| 440097
|
DBX2
|
developing brain homeobox 2
|
| 127343
|
DMBX1
|
diencephalon/mesencephalon homeobox 1
|
| 503834
|
DPRX
|
divergent-paired related homeobox
|
| 644168
|
DRGX
|
dorsal root ganglia homeobox
|
| 80712
|
ESX1
|
ESX homeobox 1
|
| 9247
|
GCM2
|
glial cells missing homolog 2
|
| 145258
|
GSC
|
goosecoid homeobox
|
| 2928
|
GSC2
|
goosecoid homeobox 2
|
| 8820
|
HESX1
|
HESX homeobox 1
|
| 84525
|
HOPX
|
HOP homeobox
|
| 26160
|
IFT172
|
intraflagellar transport 172
|
| 26525
|
IL36RN
|
interleukin 36 receptor antagonist
|
| 3670
|
ISL1
|
ISL LIM homeobox 1
|
| 64843
|
ISL2
|
ISL LIM homeobox 2
|
| 91464
|
ISX
|
intestine specific homeobox
|
| 10660
|
LBX1
|
ladybird homeobox 1
|
| 85474
|
LBX2
|
ladybird homeobox 2
|
| 9079
|
LDB1
|
LIM domain binding 2
|
| 9079
|
LDB2
|
LIM domain binding 2
|
| 3975
|
LHX1
|
LIM homeobox 1
|
| 9355
|
LHX2
|
LIM homeobox 2
|
| 8022
|
LHX3
|
LIM homeobox 3
|
| 89884
|
LHX4
|
LIM homeobox 4
|
| 64211
|
LHX5
|
LIM homeobox 5
|
| 26468
|
LHX6
|
LIM homeobox 6
|
| 431707
|
LHX8
|
LIM homeobox 8
|
| 56956
|
LHX9
|
LIM homeobox 9
|
| 4004
|
LMO1
|
LIM domain only 1
|
| 4005
|
LMO2
|
LIM domain only 2
|
| 55885
|
LMO3
|
LIM domain only 3
|
| 8543
|
LMO4
|
LIM domain only 4
|
| 4009
|
LMX1A
|
LIM homeobox transcription factor 1 alpha
|
| 4010
|
LMX1B
|
LIM homeobox transcription factor 1 beta
|
| 390010
|
NKX1-2
|
NK1 homeobox 2
|
| 135935
|
NOBOX
|
NOBOX oogenesis homeobox
|
| 23440
|
OTP
|
orthopedia homeobox
|
| 5015
|
OTX2
|
orthodenticle homeobox 2
|
| 5626
|
PROP1
|
PROP paired-like homeobox 1
|
| 5396
|
PRRX1
|
paired related homeobox 1
|
| 51450
|
PRRX2
|
paired related homeobox 2
|
| 30062
|
RAX
|
retina and anterior neural fold homeobox
|
| 84839
|
RAX2
|
retina and anterior neural fold homeobox 2
|
| 158800
|
RHOXF1
|
Rhox homeobox family member 1
|
| 84528
|
RHOXF2
|
Rhox homeobox family member 2
|
| 727940
|
RHOXF2B
|
Rhox homeobox family member 2B
|
| 645832
|
SEBOX
|
SEBOX homeobox
|
| 6473
|
SHOX
|
short stature homeobox
|
| 6474
|
SHOX2
|
short stature homeobox 2
|
| 23635
|
SSBP2
|
single stranded DNA binding protein 2
|
| 23648
|
SSBP3
|
single stranded DNA binding protein 3
|
| 170463
|
SSBP4
|
single stranded DNA binding protein 4
|
| 340260
|
UNCX
|
UNC homeobox
|
| 118472
|
ZNF511
|
zinc finger protein 511
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
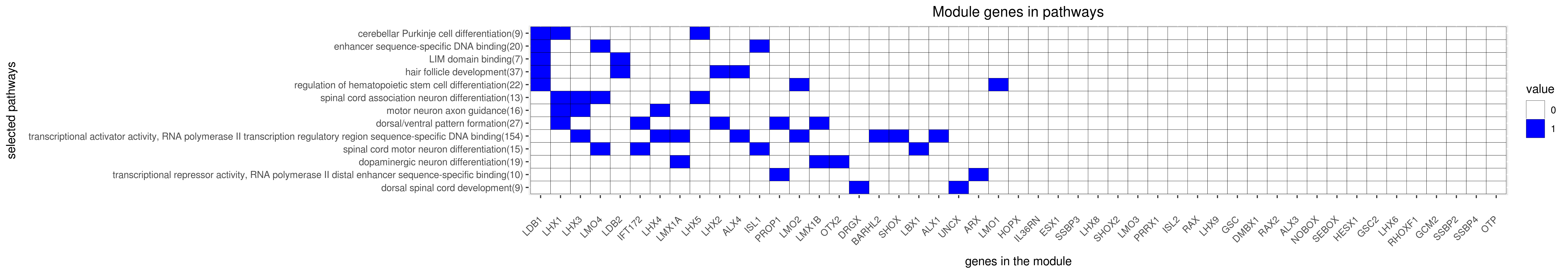
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 1.21e-03
|
1.39e-01
|
SYNTHESIS SECRETION AND INACTIVATION OF GIP
|
| 2.42e-03
|
2.51e-01
|
INCRETIN SYNTHESIS SECRETION AND INACTIVATION
|
| 3.25e-03
|
3.22e-01
|
REGULATION OF INSULIN SECRETION
|
| 7.05e-03
|
6.04e-01
|
INTEGRATION OF ENERGY METABOLISM
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:26:06 2018 - R2HTML