Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod120
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod120 |
| Module size |
29 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 148362
|
BROX
|
BRO1 domain and CAAX motif containing
|
| 5119
|
CHMP1A
|
charged multivesicular body protein 1A
|
| 57132
|
CHMP1B
|
charged multivesicular body protein 1B
|
| 27243
|
CHMP2A
|
charged multivesicular body protein 2A
|
| 25978
|
CHMP2B
|
charged multivesicular body protein 2B
|
| 51652
|
CHMP3
|
charged multivesicular body protein 3
|
| 29082
|
CHMP4A
|
charged multivesicular body protein 4A
|
| 51510
|
CHMP5
|
charged multivesicular body protein 5
|
| 91782
|
CHMP7
|
charged multivesicular body protein 7
|
| 93343
|
FAM125A
|
multivesicular body subunit 12A
|
| 89853
|
FAM125B
|
multivesicular body subunit 12B
|
| 113263
|
GLCCI1
|
glucocorticoid induced 1
|
| 9798
|
IST1
|
IST1, ESCRT-III associated factor
|
| 129531
|
MITD1
|
microtubule interacting and trafficking domain containing 1
|
| 123606
|
NIPA1
|
NIPA magnesium transporter 1
|
| 100526767
|
RNF103-CHMP3
|
RNF103-CHMP3 readthrough
|
| 11267
|
SNF8
|
SNF8, ESCRT-II complex subunit
|
| 7251
|
TSG101
|
tumor susceptibility 101
|
| 51271
|
UBAP1
|
ubiquitin associated protein 1
|
| 159195
|
USP54
|
ubiquitin specific peptidase 54
|
| 84313
|
VPS25
|
vacuolar protein sorting 25 homolog
|
| 300052
|
VPS28
|
VPS28, ESCRT-I subunit
|
| 51028
|
VPS36
|
vacuolar protein sorting 36 homolog
|
| 137492
|
VPS37A
|
VPS37A, ESCRT-I subunit
|
| 79720
|
VPS37B
|
VPS37B, ESCRT-I subunit
|
| 55048
|
VPS37C
|
VPS37C, ESCRT-I subunit
|
| 27183
|
VPS4A
|
vacuolar protein sorting 4 homolog A
|
| 9525
|
VPS4B
|
vacuolar protein sorting 4 homolog B
|
| 51534
|
VTA1
|
vesicle trafficking 1
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
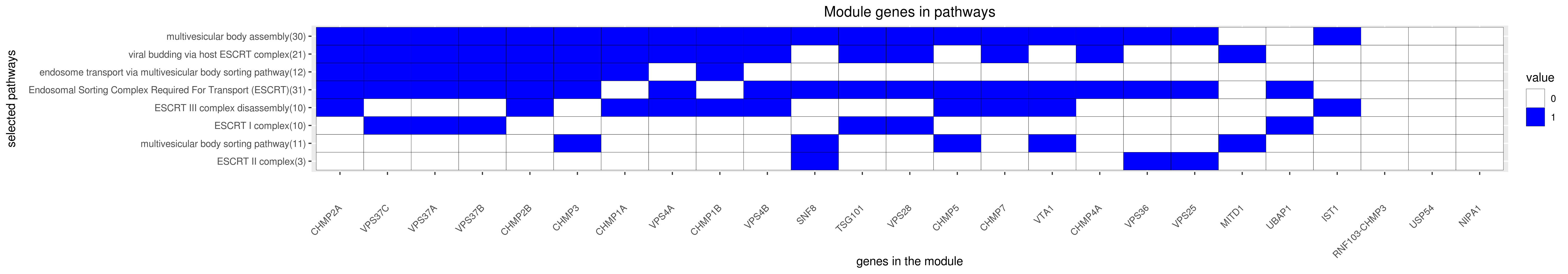
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0.00e+00
|
0.00e+00
|
ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT
|
| 0.00e+00
|
0.00e+00
|
MEMBRANE TRAFFICKING
|
| 0.00e+00
|
0.00e+00
|
ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT
|
| 0.00e+00
|
0.00e+00
|
MEMBRANE TRAFFICKING
|
| 9.47e-12
|
3.65e-09
|
MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS
|
| 5.12e-10
|
1.29e-07
|
MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS
|
| 4.19e-09
|
9.52e-07
|
LATE PHASE OF HIV LIFE CYCLE
|
| 3.99e-08
|
8.20e-06
|
HIV LIFE CYCLE
|
| 7.43e-08
|
1.95e-05
|
LATE PHASE OF HIV LIFE CYCLE
|
| 1.43e-07
|
3.63e-05
|
HIV LIFE CYCLE
|
| 3.76e-07
|
6.92e-05
|
HIV INFECTION
|
| 5.00e-07
|
1.18e-04
|
HIV INFECTION
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:25:39 2018 - R2HTML