Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod12
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod12 |
| Module size |
40 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 11149
|
BVES
|
blood vessel epicardial substance
|
| 55765
|
C1orf106
|
innate immunity activator
|
| 90060
|
CCDC120
|
coiled-coil domain containing 120
|
| 285025
|
CCDC141
|
coiled-coil domain containing 141
|
| 27185
|
DISC1
|
DISC1 scaffold protein
|
| 165215
|
FAM171B
|
family with sequence similarity 171 member B
|
| 55691
|
FRMD4A
|
FERM domain containing 4A
|
| 23150
|
FRMD4B
|
FERM domain containing 4B
|
| 392465
|
GLOD5
|
glyoxalase domain containing 5
|
| 84868
|
HAVCR2
|
hepatitis A virus cellular receptor 2
|
| 55846
|
ITFG2
|
integrin alpha FG-GAP repeat containing 2
|
| 10962
|
MLLT11
|
MLLT11, transcription factor 7 cofactor
|
| 758
|
MPPED1
|
metallophosphoesterase domain containing 1
|
| 4753
|
NELL2
|
neural EGFL like 2
|
| 22829
|
NLGN4Y
|
neuroligin 4 Y-linked
|
| 9378
|
NRXN1
|
neurexin 1
|
| 9379
|
NRXN2
|
neurexin 2
|
| 30010
|
NXPH1
|
neurexophilin 1
|
| 11249
|
NXPH2
|
neurexophilin 2
|
| 11142
|
PKIG
|
cAMP-dependent protein kinase inhibitor gamma
|
| 64091
|
POPDC2
|
popeye domain containing 2
|
| 64208
|
POPDC3
|
popeye domain containing 3
|
| 79608
|
RIC3
|
RIC3 acetylcholine receptor chaperone
|
| 79641
|
ROGDI
|
rogdi homolog
|
| 22902
|
RUFY3
|
RUN and FYVE domain containing 3
|
| 55974
|
SLC50A1
|
solute carrier family 50 member 1
|
| 6853
|
SYN1
|
synapsin I
|
| 8224
|
SYN3
|
synapsin III
|
| 341359
|
SYT10
|
synaptotagmin 10
|
| 57586
|
SYT13
|
synaptotagmin 13
|
| 255928
|
SYT14
|
synaptotagmin 14
|
| 83851
|
SYT16
|
synaptotagmin 16
|
| 148281
|
SYT6
|
synaptotagmin 6
|
| 143425
|
SYT9
|
synaptotagmin 9
|
| 83604
|
TMEM47
|
transmembrane protein 47
|
| 25789
|
TMEM59L
|
transmembrane protein 59 like
|
| 162461
|
TMEM92
|
transmembrane protein 92
|
| 7442
|
TRPV1
|
transient receptor potential cation channel subfamily V member 1
|
| 51393
|
TRPV2
|
transient receptor potential cation channel subfamily V member 2
|
| 7102
|
TSPAN7
|
tetraspanin 7
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
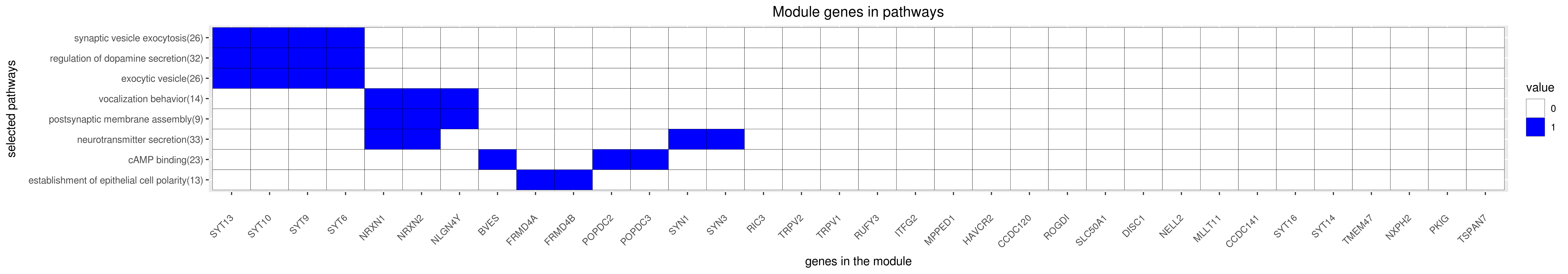
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 8.79e-06
|
1.70e-03
|
DOPAMINE NEUROTRANSMITTER RELEASE CYCLE
|
| 9.34e-06
|
1.40e-03
|
DOPAMINE NEUROTRANSMITTER RELEASE CYCLE
|
| 5.92e-05
|
9.72e-03
|
NEUROTRANSMITTER RELEASE CYCLE
|
| 6.29e-05
|
1.03e-02
|
TRANSMISSION ACROSS CHEMICAL SYNAPSES
|
| 2.01e-04
|
2.38e-02
|
NEUROTRANSMITTER RELEASE CYCLE
|
| 7.59e-04
|
7.85e-02
|
TRANSMISSION ACROSS CHEMICAL SYNAPSES
|
| 1.19e-03
|
1.17e-01
|
NEURONAL SYSTEM
|
| 5.00e-03
|
4.57e-01
|
NEURONAL SYSTEM
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:20:13 2018 - R2HTML