Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod116
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod116 |
| Module size |
21 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 11174
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif 6
|
| 1134
|
CHRNA1
|
cholinergic receptor nicotinic alpha 1 subunit
|
| 9815
|
GIT2
|
GIT ArfGAP 2
|
| 3672
|
ITGA1
|
integrin subunit alpha 1
|
| 284217
|
LAMA1
|
laminin subunit alpha 1
|
| 3908
|
LAMA2
|
laminin subunit alpha 2
|
| 3910
|
LAMA3
|
laminin subunit alpha 4
|
| 3911
|
LAMA5
|
laminin subunit alpha 5
|
| 3912
|
LAMB1
|
laminin subunit beta 1
|
| 3915
|
LAMB2
|
laminin subunit gamma 1
|
| 3914
|
LAMB3
|
laminin subunit beta 3
|
| 3915
|
LAMC1
|
laminin subunit gamma 1
|
| 3918
|
LAMC2
|
laminin subunit gamma 2
|
| 10319
|
LAMC3
|
laminin subunit gamma 3
|
| 91355
|
LRP5L
|
LDL receptor related protein 5 like
|
| 1955
|
MEGF9
|
multiple EGF like domains 9
|
| 5913
|
RAPSN
|
receptor associated protein of the synapse
|
| 9900
|
SV2A
|
synaptic vesicle glycoprotein 2A
|
| 9899
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
| 22987
|
SV2C
|
synaptic vesicle glycoprotein 2C
|
| 79875
|
THSD4
|
thrombospondin type 1 domain containing 4
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
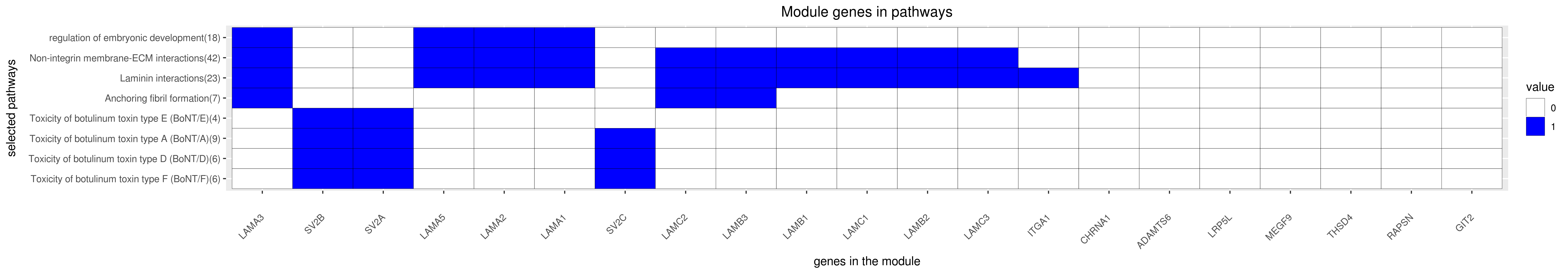
Gene membership
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 3.29e-11
|
9.41e-09
|
INTEGRIN CELL SURFACE INTERACTIONS
|
| 6.59e-08
|
1.74e-05
|
INTEGRIN CELL SURFACE INTERACTIONS
|
| 2.80e-06
|
4.53e-04
|
L1CAM INTERACTIONS
|
| 3.37e-05
|
4.64e-03
|
CELL JUNCTION ORGANIZATION
|
| 1.29e-04
|
1.95e-02
|
L1CAM INTERACTIONS
|
| 1.86e-04
|
2.22e-02
|
CELL CELL COMMUNICATION
|
| 3.56e-04
|
3.99e-02
|
AXON GUIDANCE
|
| 1.54e-03
|
1.71e-01
|
CELL JUNCTION ORGANIZATION
|
| 4.90e-03
|
4.50e-01
|
CELL CELL COMMUNICATION
|
| 8.57e-03
|
7.08e-01
|
PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS
|
| 8.57e-03
|
7.08e-01
|
HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS
|
| 8.58e-03
|
7.09e-01
|
ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS
|
| 8.61e-03
|
7.11e-01
|
AXON GUIDANCE
|
| 1.49e-02
|
1.00e+00
|
OTHER SEMAPHORIN INTERACTIONS
|
| 2.15e-02
|
1.00e+00
|
PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS
|
| 2.15e-02
|
1.00e+00
|
HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS
|
| 2.68e-02
|
1.00e+00
|
ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS
|
| 3.12e-02
|
1.00e+00
|
OTHER SEMAPHORIN INTERACTIONS
|
| 3.49e-02
|
1.00e+00
|
DEVELOPMENTAL BIOLOGY
|
| 4.23e-02
|
1.00e+00
|
SMOOTH MUSCLE CONTRACTION
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:25:27 2018 - R2HTML