Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod114
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod114 |
| Module size |
37 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 173
|
AFM
|
afamin
|
| 259
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
| 346
|
APOC4
|
apolipoprotein C4
|
| 350
|
APOH
|
apolipoprotein H
|
| 55937
|
APOM
|
apolipoprotein M
|
| 28834
|
C7
|
immunoglobulin lambda constant 7
|
| 731
|
C8A
|
complement C8 alpha chain
|
| 732
|
C8B
|
complement C8 beta chain
|
| 733
|
C8G
|
complement C8 gamma chain
|
| 1645
|
C9
|
aldo-keto reductase family 1 member C1
|
| 7123
|
CLEC3B
|
C-type lectin domain family 3 member B
|
| 1361
|
CPB2
|
carboxypeptidase B2
|
| 2165
|
F13B
|
coagulation factor XIII B chain
|
| 2147
|
F2
|
coagulation factor II, thrombin
|
| 26998
|
FETUB
|
fetuin B
|
| 2243
|
FGA
|
fibrinogen alpha chain
|
| 2244
|
FGB
|
fibrinogen beta chain
|
| 2266
|
FGG
|
fibrinogen gamma chain
|
| 2267
|
FGL1
|
fibrinogen like 1
|
| 10875
|
FGL2
|
fibrinogen like 2
|
| 2629
|
GC
|
glucosylceramidase beta
|
| 55876
|
GSDMB
|
gasdermin B
|
| 3263
|
HPX
|
hemopexin
|
| 3485
|
IGFBP2
|
insulin like growth factor binding protein 2
|
| 4018
|
LPA
|
lipoprotein(a)
|
| 11223
|
MST1L
|
macrophage stimulating 1 like
|
| 5004
|
ORM1
|
orosomucoid 1
|
| 5005
|
ORM2
|
orosomucoid 2
|
| 308942
|
PLAT
|
DENN domain containing 5A
|
| 5340
|
PLG
|
plasminogen
|
| 55848
|
PLGRKT
|
plasminogen receptor with a C-terminal lysine
|
| 5444
|
PON1
|
paraoxonase 1
|
| 8858
|
PROZ
|
protein Z, vitamin K dependent plasma glycoprotein
|
| 5858
|
PZP
|
PZP, alpha-2-macroglobulin like
|
| 462
|
SERPINC1
|
serpin family C member 1
|
| 6694
|
SPP2
|
secreted phosphoprotein 2
|
| 7448
|
VTN
|
vitronectin
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
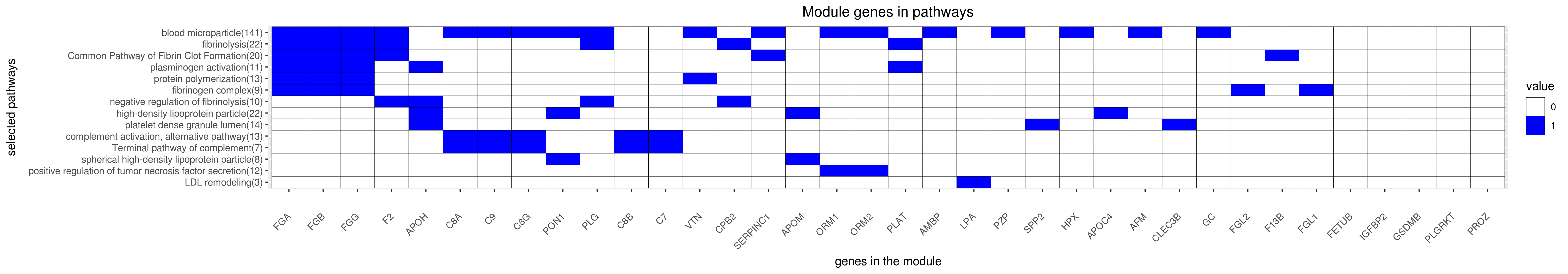
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 1.10e-12
|
3.26e-10
|
COMMON PATHWAY
|
| 1.01e-11
|
3.89e-09
|
COMMON PATHWAY
|
| 8.50e-10
|
2.09e-07
|
FORMATION OF FIBRIN CLOT CLOTTING CASCADE
|
| 1.54e-09
|
4.98e-07
|
FORMATION OF FIBRIN CLOT CLOTTING CASCADE
|
| 5.34e-09
|
1.20e-06
|
COMPLEMENT CASCADE
|
| 5.46e-08
|
1.46e-05
|
COMPLEMENT CASCADE
|
| 1.00e-05
|
1.49e-03
|
PLATELET AGGREGATION PLUG FORMATION
|
| 2.23e-05
|
3.99e-03
|
REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS
|
| 2.68e-05
|
3.75e-03
|
GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS
|
| 2.77e-05
|
3.86e-03
|
GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS
|
| 3.54e-05
|
4.86e-03
|
P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS
|
| 4.57e-05
|
7.70e-03
|
PLATELET AGGREGATION PLUG FORMATION
|
| 9.02e-05
|
1.42e-02
|
P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS
|
| 9.02e-05
|
1.42e-02
|
GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS
|
| 1.13e-04
|
1.42e-02
|
REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS
|
| 1.89e-04
|
2.25e-02
|
INTEGRIN ALPHAIIB BETA3 SIGNALING
|
| 2.56e-04
|
2.97e-02
|
INTEGRIN CELL SURFACE INTERACTIONS
|
| 2.79e-04
|
3.85e-02
|
GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS
|
| 2.87e-04
|
3.94e-02
|
RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2
|
| 3.49e-04
|
3.93e-02
|
RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:25:22 2018 - R2HTML