Back to main page
DREAM Module Identification Challenge – Consensus modules
PPI-STRING_Consensus_mod1
| Assigned name |
NA |
| Network |
PPI-STRING |
| Module ID |
PPI-STRING_Consensus_mod1 |
| Module size |
33 genes |
|
Module genes
This module comprises the following genes:
| Gene ID |
Gene Symbol |
Gene Name |
| 712
|
C1QA
|
complement C1q A chain
|
| 713
|
C1QB
|
complement C1q B chain
|
| 714
|
C1QC
|
complement C1q C chain
|
| 715
|
C1R
|
complement C1r
|
| 716
|
C1S
|
complement C1s
|
| 308846
|
C2
|
UV radiation resistance associated
|
| 720
|
C4A
|
complement C4A (Rodgers blood group)
|
| 721
|
C4B
|
complement C4B (Chido blood group)
|
| 722
|
C4BPA
|
complement component 4 binding protein alpha
|
| 725
|
C4BPB
|
complement component 4 binding protein beta
|
| 4179
|
CD46
|
CD46 molecule
|
| 975
|
CD81
|
CD81 molecule
|
| 629
|
CFB
|
complement factor B
|
| 3075
|
CFH
|
complement factor H
|
| 3078
|
CFHR1
|
complement factor H related 1
|
| 3426
|
CFI
|
complement factor I
|
| 1378
|
CR1
|
complement C3b/C4b receptor 1 (Knops blood group)
|
| 1380
|
CR2
|
complement C3d receptor 2
|
| 1465
|
CRP
|
cysteine and glycine rich protein 1
|
| 114784
|
CSMD2
|
CUB and Sushi multiple domains 2
|
| 114788
|
CSMD3
|
CUB and Sushi multiple domains 3
|
| 2210
|
FCGR1B
|
Fc fragment of IgG receptor Ib
|
| 2219
|
FCN1
|
ficolin 1
|
| 2220
|
FCN2
|
ficolin 2
|
| 8547
|
FCN3
|
ficolin 3
|
| 5648
|
MASP1
|
mannan binding lectin serine peptidase 1
|
| 10747
|
MASP2
|
mannan binding lectin serine peptidase 2
|
| 4153
|
MBL2
|
mannose binding lectin 2
|
| 4360
|
MRC1
|
mannose receptor C-type 1
|
| 5738
|
PTGFRN
|
prostaglandin F2 receptor inhibitor
|
| 5806
|
PTX3
|
pentraxin 3
|
| 6283
|
S100A12
|
S100 calcium binding protein A12
|
| 283600
|
SLC25A47
|
solute carrier family 25 member 47
|
|
Functional annotation
Modules were tested for enrichment in functional and pathway annotations using two complementary approaches:
1. To select a small number of specific / non-redundant annotations for each module, a regression-based approach was used;
2. To obtain the complete set of enriched annotations, an extension of Fisher’s exact test that takes annotation bias into account was employed (Wallenius’ non-central hypergeometric distribution).
Most specific annotations for this module
1Regression coefficient
2Fisher’s exact test nominal P-value
3Annotation source (Reactome, GO biological process (BP), molecular function (MF) and cellular component (CC))
4GO category or Reactome pathway
5High-level branch of annotation tree
Gene membership
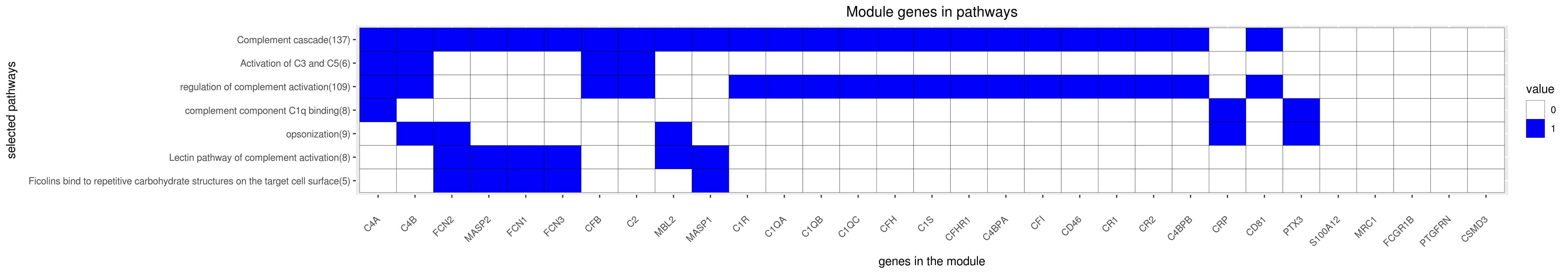
All enriched annotations
Gene Ontology
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Reactome
| P-value1 |
FDR2 |
Term |
| 0.00e+00
|
0.00e+00
|
COMPLEMENT CASCADE
|
| 0.00e+00
|
0.00e+00
|
INITIAL TRIGGERING OF COMPLEMENT
|
| 0.00e+00
|
0.00e+00
|
INNATE IMMUNE SYSTEM
|
| 0.00e+00
|
0.00e+00
|
CREATION OF C4 AND C2 ACTIVATORS
|
| 0.00e+00
|
0.00e+00
|
REGULATION OF COMPLEMENT CASCADE
|
| 0.00e+00
|
0.00e+00
|
COMPLEMENT CASCADE
|
| 0.00e+00
|
0.00e+00
|
INITIAL TRIGGERING OF COMPLEMENT
|
| 0.00e+00
|
0.00e+00
|
INNATE IMMUNE SYSTEM
|
| 0.00e+00
|
0.00e+00
|
CREATION OF C4 AND C2 ACTIVATORS
|
| 0.00e+00
|
0.00e+00
|
REGULATION OF COMPLEMENT CASCADE
|
| 1.10e-04
|
1.69e-02
|
CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES
|
| 4.05e-03
|
3.43e-01
|
CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES
|
| 5.84e-03
|
5.20e-01
|
ANTIGEN PROCESSING CROSS PRESENTATION
|
| 1.62e-02
|
1.00e+00
|
ANTIGEN PROCESSING CROSS PRESENTATION
|
| 2.05e-02
|
1.00e+00
|
TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX
|
| 2.59e-02
|
1.00e+00
|
ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING
|
| 2.89e-02
|
1.00e+00
|
ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING
|
| 3.16e-02
|
1.00e+00
|
RIP MEDIATED NFKB ACTIVATION VIA DAI
|
| 3.57e-02
|
1.00e+00
|
TRAF6 MEDIATED NFKB ACTIVATION
|
| 4.00e-02
|
1.00e+00
|
RIP MEDIATED NFKB ACTIVATION VIA DAI
|
|
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Mouse mutant phenotypes
11Nominal enrichment p-value (Wallenius’ noncentral hypergeometric distribution)
2FDR corrected p-value (Benjamini-Hochberg)
Generated on: Thu Aug 30 17:19:40 2018 - R2HTML